ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex
- PMID: 9658116
- PMCID: PMC109868
- DOI: 10.1128/JVI.72.8.6689-6698.1998
ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex
Abstract
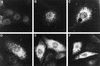
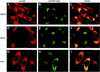
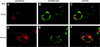
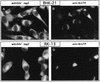
Among the functions of the replicase of equine arteritis virus (EAV; family Arteriviridae, order Nidovirales) are important viral enzyme activities such as proteases and the putative RNA polymerase and RNA helicase functions. The replicase is expressed in the form of two polyproteins (open reading frame 1a [ORF1a] and ORF1ab), which are processed into 12 nonstructural proteins by three viral proteases. In immunofluorescence assays, the majority of these cleavage products localized to the perinuclear region of the cell. A dense granular and vesicular staining was observed, which strongly suggested membrane association. By using confocal microscopy and double-label immunofluorescence, the distribution of the EAV replicase was shown to overlap with that of PDI, a resident protein of the endoplasmic reticulum and intermediate compartment. An in situ labeling of nascent viral RNA with bromo-UTP demonstrated that the membrane-bound complex in which the replicase subunits accumulate is indeed the site of viral RNA synthesis. A number of ORF1a-encoded hydrophobic domains were postulated to be involved in the membrane association of the arterivirus replication complex. By using various biochemical methods (Triton X-114 extraction, membrane purification, and sodium carbonate treatment), replicase subunits containing these domains were shown to behave as integral membrane proteins and to be membrane associated in infected cells. Thus, contribution to the formation of a membrane-bound scaffold for the viral replication-transcription complex appears to be an important novel function for the arterivirus ORF1a replicase polyprotein.
Figures


Similar articles
-
Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex.J Virol. 1999 Mar;73(3):2016-26. doi: 10.1128/JVI.73.3.2016-2026.1999. J Virol. 1999. PMID: 9971782 Free PMC article.
-
Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex.J Gen Virol. 2001 May;82(Pt 5):985-994. doi: 10.1099/0022-1317-82-5-985. J Gen Virol. 2001. PMID: 11297673
-
Adaptive Mutations in Replicase Transmembrane Subunits Can Counteract Inhibition of Equine Arteritis Virus RNA Synthesis by Cyclophilin Inhibitors.J Virol. 2019 Aug 28;93(18):e00490-19. doi: 10.1128/JVI.00490-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243130 Free PMC article.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
Viral RNA replication in association with cellular membranes.Curr Top Microbiol Immunol. 2005;285:139-73. doi: 10.1007/3-540-26764-6_5. Curr Top Microbiol Immunol. 2005. PMID: 15609503 Free PMC article. Review.
Cited by
-
De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase.J Virol. 2007 Aug;81(16):8384-95. doi: 10.1128/JVI.00564-07. Epub 2007 May 30. J Virol. 2007. PMID: 17537850 Free PMC article.
-
Secondary structure and function of the 5'-proximal region of the equine arteritis virus RNA genome.RNA. 2004 Mar;10(3):424-37. doi: 10.1261/rna.5174804. RNA. 2004. PMID: 14970388 Free PMC article.
-
Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs.PLoS Pathog. 2010 Feb 19;6(2):e1000772. doi: 10.1371/journal.ppat.1000772. PLoS Pathog. 2010. PMID: 20174607 Free PMC article.
-
Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region.J Virol. 2005 May;79(10):6312-24. doi: 10.1128/JVI.79.10.6312-6324.2005. J Virol. 2005. PMID: 15858015 Free PMC article.
-
Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis.EMBO J. 2001 Dec 17;20(24):7220-8. doi: 10.1093/emboj/20.24.7220. EMBO J. 2001. PMID: 11742998 Free PMC article.
References
-
- Bonilla P J, Hughes S A, Pinon J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV A-59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. - PubMed
-
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. - PubMed
-
- Breese S S, Jr, McCollum W H. Electron microscopic characterization of equine arteritis virus. In: Bryans J T, Gerber H, editors. Proceedings of the 2nd International Conference on Equine Infectious Diseases. S. Basel, Switzerland: Karger; 1970. pp. 133–139.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

