The mouse and human genes encoding the recognition component of the N-end rule pathway
- PMID: 9653112
- PMCID: PMC20901
- DOI: 10.1073/pnas.95.14.7898
The mouse and human genes encoding the recognition component of the N-end rule pathway
Abstract
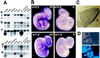
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. The N-end rule pathway is one proteolytic pathway of the ubiquitin system. The recognition component of this pathway, called N-recognin or E3, binds to a destabilizing N-terminal residue of a substrate protein and participates in the formation of a substrate-linked multiubiquitin chain. We report the cloning of the mouse and human Ubr1 cDNAs and genes that encode a mammalian N-recognin called E3alpha. Mouse UBR1p (E3alpha) is a 1,757-residue (200-kDa) protein that contains regions of sequence similarity to the 225-kDa Ubr1p of the yeast Saccharomyces cerevisiae. Mouse and human UBR1p have apparent homologs in other eukaryotes as well, thus defining a distinct family of proteins, the UBR family. The residues essential for substrate recognition by the yeast Ubr1p are conserved in the mouse UBR1p. The regions of similarity among the UBR family members include a putative zinc finger and RING-H2 finger, another zinc-binding domain. Ubr1 is located in the middle of mouse chromosome 2 and in the syntenic 15q15-q21.1 region of human chromosome 15. Mouse Ubr1 spans approximately 120 kilobases of genomic DNA and contains approximately 50 exons. Ubr1 is ubiquitously expressed in adults, with skeletal muscle and heart being the sites of highest expression. In mouse embryos, the Ubr1 expression is highest in the branchial arches and in the tail and limb buds. The cloning of Ubr1 makes possible the construction of Ubr1-lacking mouse strains, a prerequisite for the functional understanding of the mammalian N-end rule pathway.
Figures



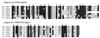

Similar articles
-
Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway.Mol Cell Biol. 2001 Dec;21(23):8007-21. doi: 10.1128/MCB.21.23.8007-8021.2001. Mol Cell Biol. 2001. PMID: 11689692 Free PMC article.
-
The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain.EMBO J. 1999 Dec 1;18(23):6832-44. doi: 10.1093/emboj/18.23.6832. EMBO J. 1999. PMID: 10581257 Free PMC article.
-
A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons.Mol Cell Biol. 2005 Aug;25(16):7120-36. doi: 10.1128/MCB.25.16.7120-7136.2005. Mol Cell Biol. 2005. PMID: 16055722 Free PMC article.
-
The N-end rule pathway of protein degradation.Genes Cells. 1997 Jan;2(1):13-28. doi: 10.1046/j.1365-2443.1997.1020301.x. Genes Cells. 1997. PMID: 9112437 Review.
-
SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions.Antioxid Redox Signal. 2001 Aug;3(4):635-50. doi: 10.1089/15230860152542989. Antioxid Redox Signal. 2001. PMID: 11554450 Review.
Cited by
-
The substrate recognition domains of the N-end rule pathway.J Biol Chem. 2009 Jan 16;284(3):1884-95. doi: 10.1074/jbc.M803641200. Epub 2008 Nov 13. J Biol Chem. 2009. PMID: 19008229 Free PMC article.
-
The ubiquitin-proteasome system in retinal health and disease.Mol Neurobiol. 2013 Apr;47(2):790-810. doi: 10.1007/s12035-012-8391-5. Epub 2013 Jan 22. Mol Neurobiol. 2013. PMID: 23339020 Review.
-
Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600.Proc Natl Acad Sci U S A. 2005 Aug 9;102(32):11492-7. doi: 10.1073/pnas.0505337102. Epub 2005 Aug 1. Proc Natl Acad Sci U S A. 2005. PMID: 16061792 Free PMC article.
-
Expression and biochemical characterization of the human enzyme N-terminal asparagine amidohydrolase.Biochemistry. 2011 Apr 12;50(14):3025-33. doi: 10.1021/bi101832w. Epub 2011 Mar 18. Biochemistry. 2011. PMID: 21375249 Free PMC article.
-
PARK7 modulates autophagic proteolysis through binding to the N-terminally arginylated form of the molecular chaperone HSPA5.Autophagy. 2018;14(11):1870-1885. doi: 10.1080/15548627.2018.1491212. Epub 2018 Jul 23. Autophagy. 2018. PMID: 29976090 Free PMC article.
References
-
- Varshavsky A. Trends Biochem Sci. 1997;22:383–387. - PubMed
-
- Hershko A. Curr Opin Cell Biol. 1997;9:788–799. - PubMed
-
- Haas A J, Siepman T J. FASEB J. 1997;11:1257–1268. - PubMed
-
- Hochstrasser M. Annu Rev Genet. 1996;30:405–439. - PubMed
-
- Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

