Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody
- PMID: 9641677
- PMCID: PMC5629912
- DOI: 10.1038/31405
Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody
Abstract
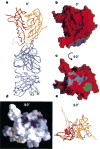
The entry of human immunodeficiency virus (HIV) into cells requires the sequential interaction of the viral exterior envelope glycoprotein, gp120, with the CD4 glycoprotein and a chemokine receptor on the cell surface. These interactions initiate a fusion of the viral and cellular membranes. Although gp120 can elicit virus-neutralizing antibodies, HIV eludes the immune system. We have solved the X-ray crystal structure at 2.5 A resolution of an HIV-1 gp120 core complexed with a two-domain fragment of human CD4 and an antigen-binding fragment of a neutralizing antibody that blocks chemokine-receptor binding. The structure reveals a cavity-laden CD4-gp120 interface, a conserved binding site for the chemokine receptor, evidence for a conformational change upon CD4 binding, the nature of a CD4-induced antibody epitope, and specific mechanisms for immune evasion. Our results provide a framework for understanding the complex biology of HIV entry into cells and should guide efforts to intervene.
Figures
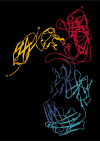



Comment in
-
HIV. Envelope's letters boxed into shape.Nature. 1998 Jun 18;393(6686):630-1. doi: 10.1038/31359. Nature. 1998. PMID: 9641673 No abstract available.
Similar articles
-
The antigenic structure of the HIV gp120 envelope glycoprotein.Nature. 1998 Jun 18;393(6686):705-11. doi: 10.1038/31514. Nature. 1998. PMID: 9641684
-
Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4.Science. 2007 Sep 28;317(5846):1930-4. doi: 10.1126/science.1145373. Science. 2007. PMID: 17901336 Free PMC article.
-
CD4 binding site antibodies inhibit human immunodeficiency virus gp120 envelope glycoprotein interaction with CCR5.J Virol. 2003 Jan;77(1):713-8. doi: 10.1128/jvi.77.1.713-718.2003. J Virol. 2003. PMID: 12477875 Free PMC article.
-
Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: conversion of a viral entry agonist to an antagonist.Acc Chem Res. 2014 Apr 15;47(4):1228-37. doi: 10.1021/ar4002735. Epub 2014 Feb 6. Acc Chem Res. 2014. PMID: 24502450 Free PMC article. Review.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
Cited by
-
Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.Cell. 2015 Jun 4;161(6):1280-92. doi: 10.1016/j.cell.2015.05.007. Epub 2015 May 21. Cell. 2015. PMID: 26004070 Free PMC article.
-
HIV-1 gp120 as a therapeutic target: navigating a moving labyrinth.Expert Opin Ther Targets. 2015 Jun;19(6):765-83. doi: 10.1517/14728222.2015.1010513. Epub 2015 Feb 27. Expert Opin Ther Targets. 2015. PMID: 25724219 Free PMC article. Review.
-
Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):6049-54. doi: 10.1073/pnas.1303682110. Epub 2013 Mar 22. Proc Natl Acad Sci U S A. 2013. PMID: 23524883 Free PMC article.
-
A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans.Immunity. 2016 Jul 19;45(1):31-45. doi: 10.1016/j.immuni.2016.06.026. Immunity. 2016. PMID: 27438765 Free PMC article.
-
Virus-receptor interactions and receptor-mediated virus entry into host cells.Subcell Biochem. 2013;68:441-66. doi: 10.1007/978-94-007-6552-8_15. Subcell Biochem. 2013. PMID: 23737061 Free PMC article. Review.
References
-
- Barre-Sinoussi F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS) Science. 1983;220:868–871. - PubMed
-
- Gallo RC, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. - PubMed
-
- Kowalski ML, et al. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. - PubMed
-
- Lu M, Blackow S, Kim P. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nature Struct Biol. 1995;2:1075–1082. - PubMed
-
- Starcich BR, et al. Identification and characterization of conserved and variable regions of the envelope gene HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

