Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment
- PMID: 9636146
- PMCID: PMC22602
- DOI: 10.1073/pnas.95.13.7316
Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment
Abstract
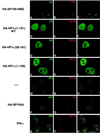
The PML/SP100 nuclear bodies (NBs) were first described as discrete subnuclear structures containing the SP100 protein. Subsequently, they were shown to contain the PML protein which is part of the oncogenic PML-RARalpha hybrid produced by the t(15;17) chromosomal translocation characteristic of acute promyelocytic leukemia. Yet, the physiological role of these nuclear bodies remains unknown. Here, we show that SP100 binds to members of the heterochromatin protein 1 (HP1) families of non-histone chromosomal proteins. Further, we demonstrate that a naturally occurring splice variant of SP100, here called SP100-HMG, is a member of the high mobility group-1 (HMG-1) protein family and may thus possess DNA-binding potential. Both HP1 and SP100-HMG concentrate in the PML/SP100 NBs, and overexpression of SP100 leads to enhanced accumulation of endogenous HP1 in these structures. When bound to a promoter, SP100, SP100-HMG and HP1 behave as transcriptional repressors in transfected mammalian cells. These observations present molecular evidence for an association between the PML/SP100 NBs and the chromatin nuclear compartment. They support a model in which the NBs may play a role in certain aspects of chromatin dynamics.
Figures

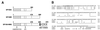


Similar articles
-
Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification.Mol Cell Biol. 2001 May;21(10):3314-24. doi: 10.1128/MCB.21.10.3314-3324.2001. Mol Cell Biol. 2001. PMID: 11313457 Free PMC article.
-
Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif.J Cell Sci. 1999 Mar;112 ( Pt 5):733-47. doi: 10.1242/jcs.112.5.733. J Cell Sci. 1999. PMID: 9973607
-
Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100.EMBO J. 2005 Oct 19;24(20):3565-75. doi: 10.1038/sj.emboj.7600820. Epub 2005 Sep 22. EMBO J. 2005. PMID: 16177824 Free PMC article.
-
PML and PML nuclear bodies: implications in antiviral defence.Biochimie. 2007 Jun-Jul;89(6-7):819-30. doi: 10.1016/j.biochi.2007.01.004. Epub 2007 Jan 27. Biochimie. 2007. PMID: 17343971 Review.
-
The Fate of Speckled Protein 100 (Sp100) During Herpesviruses Infection.Front Cell Infect Microbiol. 2021 Feb 1;10:607526. doi: 10.3389/fcimb.2020.607526. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33598438 Free PMC article. Review.
Cited by
-
Conservation of heterochromatin protein 1 function.Mol Cell Biol. 2000 Sep;20(18):6970-83. doi: 10.1128/MCB.20.18.6970-6983.2000. Mol Cell Biol. 2000. PMID: 10958692 Free PMC article.
-
Dynamic associations of heterochromatin protein 1 with the nuclear envelope.EMBO J. 2000 Dec 1;19(23):6558-68. doi: 10.1093/emboj/19.23.6558. EMBO J. 2000. PMID: 11101528 Free PMC article.
-
Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha.EMBO J. 2002 Nov 1;21(21):5797-806. doi: 10.1093/emboj/cdf560. EMBO J. 2002. PMID: 12411497 Free PMC article.
-
Human Slug is a repressor that localizes to sites of active transcription.Mol Cell Biol. 2000 Jul;20(14):5087-95. doi: 10.1128/MCB.20.14.5087-5095.2000. Mol Cell Biol. 2000. PMID: 10866665 Free PMC article.
-
Sp100A is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation.Oncogene. 2016 Jun 16;35(24):3178-89. doi: 10.1038/onc.2015.378. Epub 2015 Oct 19. Oncogene. 2016. PMID: 26477309
References
-
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Cell. 1994;76:345–356. - PubMed
-
- Maul G G, Yu E, Ishov A M, Epstein A L. J Cell Biochem. 1995;59:498–513. - PubMed
-
- Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

