A novel Myb homolog initiates Dictyostelium development by induction of adenylyl cyclase expression
- PMID: 9620859
- PMCID: PMC316870
- DOI: 10.1101/gad.12.11.1738
A novel Myb homolog initiates Dictyostelium development by induction of adenylyl cyclase expression
Abstract
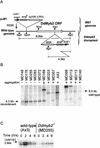
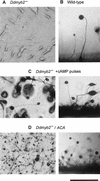
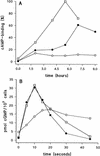
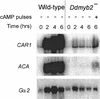
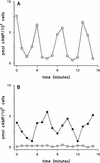
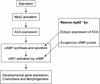
Dictyostelium development is induced by starvation. The adenylyl cyclase gene ACA is one of the first genes expressed upon starvation. ACA produces extracellular cAMP that induces chemotaxis, aggregation, and differentiation in neighboring cells. Using insertional mutagenesis we have isolated a mutant that does not aggregate upon starvation but is rescued by adding extracellular cAMP. Sequencing of the mutated locus revealed a new gene, DdMYB2, whose product contains three Myb repeats, the DNA-binding motif of Myb-related transcription factors. Ddmyb2-null cells show undetectable levels of ACA transcript and no cAMP production. Ectopic expression of ACA from a constitutive promotor rescues differentiation and morphogenesis of Ddmyb2-null mutants. The results suggest that development in Dictyostelium starts by starvation-mediated DdMyb2 activation, which induces adenylyl cyclase activity producing the differentiation-inducing signal cAMP.
Figures



Similar articles
-
amiB, a novel gene required for the growth/differentiation transition in Dictyostelium.Genes Cells. 2000 Jan;5(1):43-55. doi: 10.1046/j.1365-2443.2000.00305.x. Genes Cells. 2000. PMID: 10651904
-
A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium.Mol Biol Cell. 1999 Sep;10(9):2829-45. doi: 10.1091/mbc.10.9.2829. Mol Biol Cell. 1999. PMID: 10473630 Free PMC article.
-
Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium.Dev Biol. 1997 Mar 15;183(2):208-21. doi: 10.1006/dbio.1996.8499. Dev Biol. 1997. PMID: 9126295
-
Adenylyl cyclase expression and regulation during the differentiation of Dictyostelium discoideum.IUBMB Life. 2004 Sep;56(9):541-6. doi: 10.1080/15216540400013887. IUBMB Life. 2004. PMID: 15590560 Review.
-
Dispatch. Dictyostelium chemotaxis: fascism through the back door?Curr Biol. 2003 Apr 29;13(9):R353-4. doi: 10.1016/s0960-9822(03)00274-4. Curr Biol. 2003. PMID: 12725750 Review.
Cited by
-
Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer.Science. 2014 Mar 21;343(6177):1249531. doi: 10.1126/science.1249531. Science. 2014. PMID: 24653039 Free PMC article.
-
Genome-Wide Classification of Myb Domain-Containing Protein Families in Entamoeba invadens.Genes (Basel). 2024 Feb 2;15(2):201. doi: 10.3390/genes15020201. Genes (Basel). 2024. PMID: 38397191 Free PMC article.
-
Independent Evolution of the MYB Family in Brown Algae.Front Genet. 2022 Feb 4;12:811993. doi: 10.3389/fgene.2021.811993. eCollection 2021. Front Genet. 2022. PMID: 35186015 Free PMC article.
-
cAMP signaling in Dictyostelium. Complexity of cAMP synthesis, degradation and detection.J Muscle Res Cell Motil. 2002;23(7-8):793-802. doi: 10.1023/a:1024483829878. J Muscle Res Cell Motil. 2002. PMID: 12952077 Review.
-
CbfA, the C-module DNA-binding factor, plays an essential role in the initiation of Dictyostelium discoideum development.Eukaryot Cell. 2004 Oct;3(5):1349-58. doi: 10.1128/EC.3.5.1349-1358.2004. Eukaryot Cell. 2004. PMID: 15470262 Free PMC article.
References
-
- Amaravadi L, King MW. Characterization and expression of the Xenopus c-Myb homolog. Oncogene. 1994;9:971–974. - PubMed
-
- Clark KA, McKearin DM. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development. 1996;122:937–950. - PubMed
-
- Frampton J, Leutz A, Gibson TJ, Graf T. DNA-binding domain ancestry. Nature. 1989;342:134. - PubMed
-
- Garcia-Bustos J, Heitman J, Hall MN. Nuclear protein localisation. Biochim Biophys Acta. 1991;1071:83–101. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
