An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription
- PMID: 9618479
- PMCID: PMC22611
- DOI: 10.1073/pnas.95.12.6722
An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription
Abstract
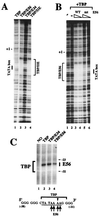
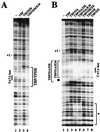
The basal transcription factor IIE (TFIIE) is thought to be one of the last factors to be assembled into a preinitiation complex (PIC) at eukaryotic promoters after RNA polymerase II and TFIIF have been incorporated. It was shown that a primary function of TFIIE is to recruit and cooperate with TFIIH in promoter melting. Here, we show that the large subunit of TFIIE (E56) can directly stimulate TBP binding to the promoter in the absence of other basal factors. The zinc-finger domain of E56, required for transcriptional activity, is critical for this function. In addition, the small subunit of TFIIE (E34) directly contacts DNA and TFIIA and thus providing a second mechanism for TFIIE to help binding of a TBP/IIA complex to the promoter, the first critical step in the PIC assembly. These studies suggest an alternative PIC assembly pathway in which TFIIE affects both TBP and TFIIH functions during initiation of RNA synthesis.
Figures

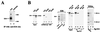
Similar articles
-
Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance.Genes Dev. 1994 Mar 1;8(5):515-24. doi: 10.1101/gad.8.5.515. Genes Dev. 1994. PMID: 7926747
-
Transcriptional activity of transcription factor IIE is dependent on zinc binding.Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9529-33. doi: 10.1073/pnas.91.20.9529. Proc Natl Acad Sci U S A. 1994. PMID: 7937800 Free PMC article.
-
Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II.Genes Dev. 1993 Jul;7(7A):1254-65. doi: 10.1101/gad.7.7a.1254. Genes Dev. 1993. PMID: 8319911
-
Structural insights into assembly of transcription preinitiation complex.Curr Opin Struct Biol. 2022 Aug;75:102404. doi: 10.1016/j.sbi.2022.102404. Epub 2022 Jun 11. Curr Opin Struct Biol. 2022. PMID: 35700575 Review.
-
TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes.Trends Biochem Sci. 2000 Feb;25(2):59-63. doi: 10.1016/s0968-0004(99)01527-3. Trends Biochem Sci. 2000. PMID: 10664584 Review.
Cited by
-
Structural basis of transcription initiation by RNA polymerase II.Nat Rev Mol Cell Biol. 2015 Mar;16(3):129-43. doi: 10.1038/nrm3952. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693126 Review.
-
Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle.Biomolecules. 2024 Feb 1;14(2):176. doi: 10.3390/biom14020176. Biomolecules. 2024. PMID: 38397413 Free PMC article. Review.
-
How eukaryotic genes are transcribed.Crit Rev Biochem Mol Biol. 2009 Jun;44(2-3):117-41. doi: 10.1080/10409230902858785. Crit Rev Biochem Mol Biol. 2009. PMID: 19514890 Free PMC article. Review.
-
Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.EMBO J. 2000 Mar 15;19(6):1346-56. doi: 10.1093/emboj/19.6.1346. EMBO J. 2000. PMID: 10716934 Free PMC article.
-
Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock.Genes Dev. 2006 Aug 15;20(16):2250-65. doi: 10.1101/gad.1437506. Genes Dev. 2006. PMID: 16912275 Free PMC article.
References
-
- Kaiser K, Meisterernst M. Trends Biochem Sci. 1996;21:342–345. - PubMed
-
- Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. - PubMed
-
- Tjian R, Maniatis T. Cell. 1994;77:5–8. - PubMed
-
- Roeder R G. Trends Biochem Sci. 1996;21:327–335. - PubMed
-
- Dynlacht B D, Hoey T, Tjian R. Cell. 1991;55:563–576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

