Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae
- PMID: 9614172
- PMCID: PMC25348
- DOI: 10.1091/mbc.9.6.1253
Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae
Abstract
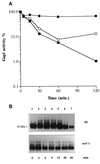
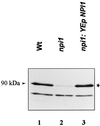
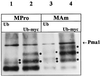
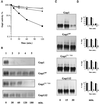
Addition of ammonium ions to yeast cells growing on proline as the sole nitrogen source induces rapid inactivation and degradation of the general amino acid permease Gap1 through a process requiring the Npi1/Rsp5 ubiquitin (Ub) ligase. In this study, we show that NH4+ induces endocytosis of Gap1, which is then delivered into the vacuole where it is degraded. This down-regulation is accompanied by increased conversion of Gap1 to ubiquitinated forms. Ubiquitination and subsequent degradation of Gap1 are impaired in the npi1 strain. In this mutant, the amount of Npi1/Rsp5 Ub ligase is reduced >10-fold compared with wild-type cells. The C-terminal tail of Gap1 contains sequences, including a di-leucine motif, which are required for NH4+-induced internalization and degradation of the permease. We show here that mutant Gap1 permeases affected in these sequences still bind Ub. Furthermore, we provide evidence that only a small fraction of Gap1 is modified by Ub after addition of NH4+ to mutants defective in endocytosis.
Figures

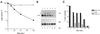
Similar articles
-
The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the gap1 permease.Biochem Biophys Res Commun. 1999 Apr 13;257(2):561-6. doi: 10.1006/bbrc.1999.0505. Biochem Biophys Res Commun. 1999. PMID: 10198251
-
NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase.Mol Microbiol. 1995 Oct;18(1):77-87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. Mol Microbiol. 1995. PMID: 8596462
-
Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1.J Biol Chem. 2001 Nov 23;276(47):43949-57. doi: 10.1074/jbc.M102945200. Epub 2001 Aug 10. J Biol Chem. 2001. PMID: 11500494
-
The ubiquitin code of yeast permease trafficking.Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
-
Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins.FASEB J. 1997 Dec;11(14):1215-26. doi: 10.1096/fasebj.11.14.9409540. FASEB J. 1997. PMID: 9409540 Review.
Cited by
-
Sul1 and Sul2 sulfate transceptors signal to protein kinase A upon exit of sulfur starvation.J Biol Chem. 2015 Apr 17;290(16):10430-46. doi: 10.1074/jbc.M114.629022. Epub 2015 Feb 27. J Biol Chem. 2015. PMID: 25724649 Free PMC article.
-
Amino acid transport through the Saccharomyces cerevisiae Gap1 permease is controlled by the Ras/cAMP pathway.Int J Biochem Cell Biol. 2008;40(3):496-502. doi: 10.1016/j.biocel.2007.08.012. Epub 2007 Aug 30. Int J Biochem Cell Biol. 2008. PMID: 17919965 Free PMC article.
-
From transporter to transceptor: signaling from transporters provokes re-evaluation of complex trafficking and regulatory controls: endocytic internalization and intracellular trafficking of nutrient transceptors may, at least in part, be governed by their signaling function.Bioessays. 2011 Nov;33(11):870-9. doi: 10.1002/bies.201100100. Epub 2011 Sep 13. Bioessays. 2011. PMID: 21913212 Free PMC article.
-
Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity.J Biol Chem. 2011 Aug 19;286(33):29376-29387. doi: 10.1074/jbc.M111.233346. Epub 2011 Jun 27. J Biol Chem. 2011. PMID: 21708947 Free PMC article.
-
Genetic Evidence for the Role of the Vacuole in Supplying Secretory Organelles with Ca2+ in Hansenula polymorpha.PLoS One. 2015 Dec 30;10(12):e0145915. doi: 10.1371/journal.pone.0145915. eCollection 2015. PLoS One. 2015. PMID: 26717478 Free PMC article.
References
-
- Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995.
-
- Béchet J, Grenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. - PubMed
-
- Bonneaud N, Ozier Kalogeropoulos O, Li GY, Labouesse M, Minvielle Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

