Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice
- PMID: 9600904
- PMCID: PMC27572
- DOI: 10.1073/pnas.95.11.5987
Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice
Abstract
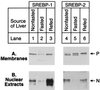
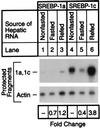
Hepatic lipid synthesis is known to be regulated by food consumption. In rodents fasting decreases the synthesis of cholesterol as well as fatty acids. Refeeding a high carbohydrate/low fat diet enhances fatty acid synthesis by 5- to 20-fold above the fed state, whereas cholesterol synthesis returns only to the prefasted level. Sterol regulatory element binding proteins (SREBPs) are transcription factors that regulate genes involved in cholesterol and fatty acid synthesis. Here, we show that fasting markedly reduces the amounts of SREBP-1 and -2 in mouse liver nuclei, with corresponding decreases in the mRNAs for SREBP-activated target genes. Refeeding a high carbohydrate/low fat diet resulted in a 4- to 5-fold increase of nuclear SREBP-1 above nonfasted levels, whereas nuclear SREBP-2 protein returned only to the nonfasted level. The hepatic mRNAs for fatty acid biosynthetic enzymes increased 5- to 10-fold above nonfasted levels, a pattern that paralleled the changes in nuclear SREBP-1. The hepatic mRNAs for enzymes involved in cholesterol synthesis returned to the nonfasted level, closely following the pattern of nuclear SREBP-2 regulation. Transgenic mice that overproduce nuclear SREBP-1c failed to show the normal decrease in hepatic mRNA levels for cholesterol and fatty acid synthetic enzymes upon fasting. We conclude that SREBPs are regulated by food consumption in the mouse liver and that the decline in nuclear SREBP-1c upon fasting may explain in part the decrease in mRNAs encoding enzymes of the fatty acid biosynthetic pathway.
Figures


Similar articles
-
Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes.J Biol Chem. 1999 Dec 10;274(50):35832-9. doi: 10.1074/jbc.274.50.35832. J Biol Chem. 1999. PMID: 10585467
-
Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c.J Biol Chem. 2002 Mar 15;277(11):9520-8. doi: 10.1074/jbc.M111421200. Epub 2002 Jan 8. J Biol Chem. 2002. PMID: 11782483
-
Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene.J Clin Invest. 1997 Oct 15;100(8):2115-24. doi: 10.1172/JCI119746. J Clin Invest. 1997. PMID: 9329978 Free PMC article.
-
Sterol regulatory element-binding proteins: activators of cholesterol and fatty acid biosynthesis.Curr Opin Lipidol. 1999 Apr;10(2):143-50. doi: 10.1097/00041433-199904000-00008. Curr Opin Lipidol. 1999. PMID: 10327282 Review.
-
Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism.Vitam Horm. 2002;65:167-94. doi: 10.1016/s0083-6729(02)65064-2. Vitam Horm. 2002. PMID: 12481547 Review.
Cited by
-
Tumor-Induced Hyperlipidemia Contributes to Tumor Growth.Cell Rep. 2016 Apr 12;15(2):336-48. doi: 10.1016/j.celrep.2016.03.020. Epub 2016 Mar 31. Cell Rep. 2016. PMID: 27050512 Free PMC article.
-
The Roles of White Adipose Tissue and Liver NADPH in Dietary Restriction-Induced Longevity.Antioxidants (Basel). 2024 Jul 8;13(7):820. doi: 10.3390/antiox13070820. Antioxidants (Basel). 2024. PMID: 39061889 Free PMC article. Review.
-
Polyunsaturated fatty acid suppression of fatty acid synthase (FASN): evidence for dietary modulation of NF-Y binding to the Fasn promoter by SREBP-1c.Biochem J. 2007 Mar 15;402(3):591-600. doi: 10.1042/BJ20061722. Biochem J. 2007. PMID: 17313375 Free PMC article.
-
Regulation by carbohydrate and clofibric acid of palmitoyl-CoA chain elongation in the liver of rats.Lipids. 2003 May;38(5):531-7. doi: 10.1007/s11745-003-1338-8. Lipids. 2003. PMID: 12880109
-
Adaptation of Oxidative Phosphorylation Machinery Compensates for Hepatic Lipotoxicity in Early Stages of MAFLD.Int J Mol Sci. 2022 Jun 20;23(12):6873. doi: 10.3390/ijms23126873. Int J Mol Sci. 2022. PMID: 35743314 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

