Optimizing the stability of single-chain proteins by linker length and composition mutagenesis
- PMID: 9600894
- PMCID: PMC34497
- DOI: 10.1073/pnas.95.11.5929
Optimizing the stability of single-chain proteins by linker length and composition mutagenesis
Abstract
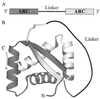
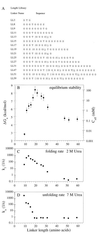
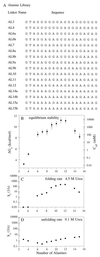
Linker length and composition were varied in libraries of single-chain Arc repressor, resulting in proteins with effective concentrations ranging over six orders of magnitude (10 microM-10 M). Linkers of 11 residues or more were required for biological activity. Equilibrium stability varied substantially with linker length, reaching a maximum for glycine-rich linkers containing 19 residues. The effects of linker length on equilibrium stability arise from significant and sometimes opposing changes in folding and unfolding kinetics. By fixing the linker length at 19 residues and varying the ratio of Ala/Gly or Ser/Gly in a 16-residue-randomized region, the effects of linker flexibility were examined. In these libraries, composition rather than sequence appears to determine stability. Maximum stability in the Ala/Gly library was observed for a protein containing 11 alanines and five glycines in the randomized region of the linker. In the Ser/Gly library, the most stable protein had seven serines and nine glycines in this region. Analysis of folding and unfolding rates suggests that alanine acts largely by accelerating folding, whereas serine acts predominantly to slow unfolding. These results demonstrate an important role for linker design in determining the stability and folding kinetics of single-chain proteins and suggest strategies for optimizing these parameters.
Figures


Similar articles
-
Effect of linker flexibility and length on the functionality of a cytotoxic engineered antibody fragment.J Biotechnol. 2015 Apr 10;199:90-7. doi: 10.1016/j.jbiotec.2015.02.008. Epub 2015 Feb 16. J Biotechnol. 2015. PMID: 25697559
-
Using loop length variants to dissect the folding pathway of a four-helix-bundle protein.J Mol Biol. 1999 Feb 12;286(1):257-65. doi: 10.1006/jmbi.1998.2474. J Mol Biol. 1999. PMID: 9931264
-
Understanding and applications of Ser/Gly linkers in protein engineering.Methods Enzymol. 2021;647:1-22. doi: 10.1016/bs.mie.2020.12.001. Epub 2020 Dec 26. Methods Enzymol. 2021. PMID: 33482985
-
Linkers: A synergistic way for the synthesis of chimeric proteins.Protein Expr Purif. 2022 Mar;191:106012. doi: 10.1016/j.pep.2021.106012. Epub 2021 Nov 10. Protein Expr Purif. 2022. PMID: 34767950 Review.
-
The analysis of protein folding kinetic data produced in protein engineering experiments.Methods. 2004 Sep;34(1):41-50. doi: 10.1016/j.ymeth.2004.03.013. Methods. 2004. PMID: 15283914 Review.
Cited by
-
Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method).Nucleic Acids Res. 2001 Aug 1;29(15):E73-3. doi: 10.1093/nar/29.15.e73. Nucleic Acids Res. 2001. PMID: 11470888 Free PMC article.
-
A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation.J Mol Biol. 2010 Mar 12;396(5):1346-60. doi: 10.1016/j.jmb.2009.12.059. Epub 2010 Jan 11. J Mol Biol. 2010. PMID: 20060838 Free PMC article.
-
A Japanese encephalitis virus peptide present on Johnson grass mosaic virus-like particles induces virus-neutralizing antibodies and protects mice against lethal challenge.J Virol. 2003 Mar;77(6):3487-94. doi: 10.1128/jvi.77.6.3487-3494.2003. J Virol. 2003. PMID: 12610124 Free PMC article.
-
Novel recombinant multiepitope proteins for the diagnosis of asymptomatic leishmania infantum-infected dogs.PLoS Negl Trop Dis. 2015 Jan 8;9(1):e3429. doi: 10.1371/journal.pntd.0003429. eCollection 2015 Jan. PLoS Negl Trop Dis. 2015. PMID: 25569685 Free PMC article.
-
Synthetic pathway for production of five-carbon alcohols from isopentenyl diphosphate.Appl Environ Microbiol. 2012 Nov;78(22):7849-55. doi: 10.1128/AEM.01175-12. Epub 2012 Aug 31. Appl Environ Microbiol. 2012. PMID: 22941086 Free PMC article.
References
-
- Bird R E, Hardman K D, Jacobson J W, Johnson S, Kaufman B M, Lee S-M, Lee T, Pope S H, Riordan G S, Whitlow M. Science. 1988;242:423–426. - PubMed
-
- Pomerantz J L, Sharp P A, Pabo C O. Science. 1995;267:93–96. - PubMed
-
- Predki P F, Regan L. Biochem. 1995;34:9834–9839. - PubMed
-
- Hallewell R A, Laria I, Tabrizi A, Carlin C, Getzoff E D, Tainer J A, Cousens L S, Mullenbach G T. J Biol Chem. 1989;264:5260–5268. - PubMed
-
- Bizub D, Weber I T, Cameron C E, Leis J P, Skalka A M. J Biol Chem. 1991;266:4951–4958. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

