Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro
- PMID: 9557686
- PMCID: PMC109626
- DOI: 10.1128/JVI.72.5.3991-3998.1998
Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro
Abstract
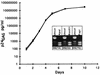
Recombinant forms of human immunodeficiency virus type 1 (HIV-1) have been shown to be of major importance in the global AIDS pandemic. Viral RNA dimer formation mediated by the dimerization initiation sequence (DIS) is believed to be essential for viral genomic RNA packaging and therefore for RNA recombination. Here, we demonstrate that HIV-1 recombination and replication are not restricted by variant DIS loop sequences. Three DIS loop forms found among HIV-1 isolates, DIS (CG), DIS (TA), and DIS (TG), when introduced into deletion mutants of HIV-1 recombined efficiently, and the progeny virions replicated with comparable kinetics. A fourth DIS loop form, containing an artificial AAAAAA sequence disrupting the putative DIS loop-loop interactions [DIS (A6)], supported efficient recombination with DIS loop variants; however, DIS (A6) progeny virions exhibited a modest replication disadvantage in mixed cultures. Our studies indicate that the nonhomologous DIS sequences found in different HIV-1 subtypes are not a primary obstacle to intersubtype recombination.
Figures

Similar articles
-
A short sequence motif in the 5' leader of the HIV-1 genome modulates extended RNA dimer formation and virus replication.J Biol Chem. 2014 Dec 19;289(51):35061-74. doi: 10.1074/jbc.M114.621425. Epub 2014 Nov 3. J Biol Chem. 2014. PMID: 25368321 Free PMC article.
-
The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells.J Virol. 2003 Aug;77(15):8329-35. doi: 10.1128/jvi.77.15.8329-8335.2003. J Virol. 2003. PMID: 12857902 Free PMC article.
-
Role of the DIS hairpin in replication of human immunodeficiency virus type 1.J Virol. 1996 Oct;70(10):6723-32. doi: 10.1128/JVI.70.10.6723-6732.1996. J Virol. 1996. PMID: 8794309 Free PMC article.
-
Identification of a major restriction in HIV-1 intersubtype recombination.Proc Natl Acad Sci U S A. 2005 Jun 21;102(25):9002-7. doi: 10.1073/pnas.0502522102. Epub 2005 Jun 14. Proc Natl Acad Sci U S A. 2005. PMID: 15956186 Free PMC article.
-
A structure-based approach for targeting the HIV-1 genomic RNA dimerization initiation site.Biochimie. 2007 Oct;89(10):1195-203. doi: 10.1016/j.biochi.2007.03.003. Epub 2007 Mar 12. Biochimie. 2007. PMID: 17434658 Review.
Cited by
-
Multiple barriers to recombination between divergent HIV-1 variants revealed by a dual-marker recombination assay.J Mol Biol. 2011 Apr 8;407(4):521-31. doi: 10.1016/j.jmb.2011.01.052. Epub 2011 Feb 3. J Mol Biol. 2011. PMID: 21295586 Free PMC article.
-
HIV-1 reverse transcription.Cold Spring Harb Perspect Med. 2012 Oct 1;2(10):a006882. doi: 10.1101/cshperspect.a006882. Cold Spring Harb Perspect Med. 2012. PMID: 23028129 Free PMC article. Review.
-
Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination.J Virol. 2007 Apr;81(8):4002-11. doi: 10.1128/JVI.02589-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267488 Free PMC article.
-
Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates.J Virol. 2003 Apr;77(8):4577-87. doi: 10.1128/jvi.77.8.4577-4587.2003. J Virol. 2003. PMID: 12663764 Free PMC article.
-
Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector.J Virol. 2002 Aug;76(15):7897-902. doi: 10.1128/jvi.76.15.7897-7902.2002. J Virol. 2002. PMID: 12097604 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

