Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection
- PMID: 9525587
- PMCID: PMC109712
- DOI: 10.1128/JVI.72.4.2697-2707.1998
Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection
Abstract
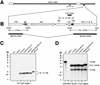
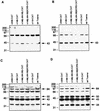
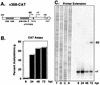
The human cytomegalovirus (HCMV) UL112-113 promoter represents a useful model for studying temporal regulation of viral gene expression. Stimulation of this promoter by the 86-kDa immediate-early protein (IE86) is controlled by sequences between nucleotides -113 and -59, which include both an ATF/CREB and an IE86 binding site. In transient assays, the ATF/CREB site is essential, and the IE86 site, although nonessential, can enhance transcription. With recombinant viruses, we have assessed the function of these promoter elements in the context of the viral genome. Transcription from the inserted UL112-113 promoter shows the same temporal pattern as the endogenous promoter, including the switch to an upstream RNA start site late in infection. Deletion of sequences containing the IE86 site results in a decrease in the level of early transcription and elimination of late transcription. In contrast, when the ATF/CREB site is deleted, early RNA synthesis is almost completely abolished, but late transcription is comparable to that of the wild type, with repositioning of the RNA start site downstream by the number of nucleotides deleted. Replacement of sequences between -108 and -95 with the HCMV cis-repression signal from the major immediate-early promoter had no effect on the level of late RNAs but resulted in the repositioning of the RNA start site 39 nucleotides upstream. These results suggest that the ATF/CREB site is functional only at early times, while sequences containing the IE86 site modulate the level of early RNAs and may be required for activating late transcription in a distance-dependent manner.
Figures

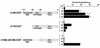



Similar articles
-
Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB.J Virol. 1995 Oct;69(10):6030-7. doi: 10.1128/JVI.69.10.6030-6037.1995. J Virol. 1995. PMID: 7666507 Free PMC article.
-
A cis repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection.J Virol. 1998 Dec;72(12):9575-84. doi: 10.1128/JVI.72.12.9575-9584.1998. J Virol. 1998. PMID: 9811691 Free PMC article.
-
Transcriptional regulation of the human cytomegalovirus US11 early gene.J Virol. 1999 Feb;73(2):863-70. doi: 10.1128/JVI.73.2.863-870.1999. J Virol. 1999. PMID: 9882286 Free PMC article.
-
Activation and regulation of human cytomegalovirus early genes.Intervirology. 1996;39(5-6):361-77. doi: 10.1159/000150507. Intervirology. 1996. PMID: 9130046 Review.
-
Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate.Clin Microbiol Rev. 1999 Jul;12(3):367-82. doi: 10.1128/CMR.12.3.367. Clin Microbiol Rev. 1999. PMID: 10398670 Free PMC article. Review.
Cited by
-
Connection between elastin haploinsufficiency and increased cell proliferation in patients with supravalvular aortic stenosis and Williams-Beuren syndrome.Am J Hum Genet. 2002 Jul;71(1):30-44. doi: 10.1086/341035. Epub 2002 May 6. Am J Hum Genet. 2002. PMID: 12016585 Free PMC article.
-
Functional properties of the human cytomegalovirus IE86 protein required for transcriptional regulation and virus replication.J Virol. 2010 Sep;84(17):8839-48. doi: 10.1128/JVI.00327-10. Epub 2010 Jun 16. J Virol. 2010. PMID: 20554773 Free PMC article.
-
Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt.Virology. 2012 Mar 15;424(2):106-14. doi: 10.1016/j.virol.2011.12.010. Epub 2012 Jan 10. Virology. 2012. PMID: 22236369 Free PMC article.
-
Novel immediate-early protein IE19 of human cytomegalovirus activates the origin recognition complex I promoter in a cooperative manner with IE72.J Virol. 2002 Apr;76(7):3158-67. doi: 10.1128/jvi.76.7.3158-3167.2002. J Virol. 2002. PMID: 11884540 Free PMC article.
-
High-throughput screening of a GlaxoSmithKline protein kinase inhibitor set identifies an inhibitor of human cytomegalovirus replication that prevents CREB and histone H3 post-translational modification.J Gen Virol. 2017 Apr;98(4):754-768. doi: 10.1099/jgv.0.000713. Epub 2017 Apr 20. J Gen Virol. 2017. PMID: 28100301 Free PMC article.
References
-
- Britt W, Alford C. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523.
-
- Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. - PubMed
-
- DeMarchi J M. Human cytomegalovirus DNA: restriction enzyme cleavage maps and map locations for immediate-early, early, and late RNAs. Virology. 1981;114:23–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

