A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition
- PMID: 9512519
- PMCID: PMC316625
- DOI: 10.1101/gad.12.6.858
A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition
Abstract
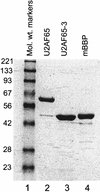
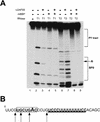
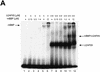
During the early events of pre-mRNA splicing, intronic cis-acting sequences are recognized and interact through a network of RNA-RNA, RNA-protein, and protein-protein contacts. Recently, we identified a branchpoint sequence binding protein in yeast (BBP). The mammalian ortholog (mBBP/SF1) also binds specifically to branchpoint sequences and interacts with the well studied mammalian splicing factor U2AF65, which binds to the adjacent polypyrimidine (PY) tract. In this paper we demonstrate that the mBBP/SF1-U2AF65 interaction promotes cooperative binding to a branchpoint sequence-polypyrimidine tract-containing RNA, and we suggest that this cooperative RNA binding contributes to initial recognition of the branchpoint sequence (BPS) during pre-mRNA splicing. We also demonstrate the essential nature of the third RBD of U2AF65 for the interaction between the two proteins, both in the presence and absence of RNA.
Figures













Similar articles
-
Recognition of RNA branch point sequences by the KH domain of splicing factor 1 (mammalian branch point binding protein) in a splicing factor complex.Mol Cell Biol. 2001 Aug;21(15):5232-41. doi: 10.1128/MCB.21.15.5232-5241.2001. Mol Cell Biol. 2001. PMID: 11438677 Free PMC article.
-
Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF.J Biol Chem. 2000 Dec 1;275(48):38059-66. doi: 10.1074/jbc.M001483200. J Biol Chem. 2000. PMID: 10954700
-
A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast.Nucleic Acids Res. 2008 May;36(8):2787-98. doi: 10.1093/nar/gkn144. Epub 2008 Mar 29. Nucleic Acids Res. 2008. PMID: 18375978 Free PMC article.
-
Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors.J Mol Biol. 2006 Feb 24;356(3):664-83. doi: 10.1016/j.jmb.2005.11.067. Epub 2005 Dec 7. J Mol Biol. 2006. PMID: 16376933 Free PMC article.
-
The branchpoint binding protein: in and out of the spliceosome cycle.Adv Exp Med Biol. 2010;693:123-41. Adv Exp Med Biol. 2010. PMID: 21189690 Review.
Cited by
-
Variations of intronic branchpoint motif: identification and functional implications in splicing and disease.Commun Biol. 2023 Nov 10;6(1):1142. doi: 10.1038/s42003-023-05513-7. Commun Biol. 2023. PMID: 37949953 Free PMC article. Review.
-
Broad variation in response of individual introns to splicing inhibitors in a humanized yeast strain.RNA. 2024 Jan 16;30(2):149-170. doi: 10.1261/rna.079866.123. RNA. 2024. PMID: 38071476 Free PMC article.
-
Disease-Causing Mutations in SF3B1 Alter Splicing by Disrupting Interaction with SUGP1.Mol Cell. 2019 Oct 3;76(1):82-95.e7. doi: 10.1016/j.molcel.2019.07.017. Epub 2019 Aug 29. Mol Cell. 2019. PMID: 31474574 Free PMC article.
-
High-affinity consensus binding of target RNAs by the STAR/GSG proteins GLD-1, STAR-2 and Quaking.BMC Mol Biol. 2010 Jun 23;11:48. doi: 10.1186/1471-2199-11-48. BMC Mol Biol. 2010. PMID: 20573244 Free PMC article.
-
Phosphorylation by SR kinases regulates the binding of PTB-associated splicing factor (PSF) to the pre-mRNA polypyrimidine tract.FEBS Lett. 2007 Jan 23;581(2):223-32. doi: 10.1016/j.febslet.2006.12.015. Epub 2006 Dec 14. FEBS Lett. 2007. PMID: 17188683 Free PMC article.
References
-
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
-
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
