Overexpression of C/EBPbeta represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein
- PMID: 9499067
- PMCID: PMC109506
- DOI: 10.1128/JVI.72.3.2113-2124.1998
Overexpression of C/EBPbeta represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein
Abstract
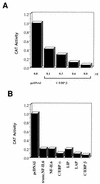
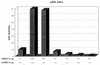
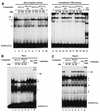
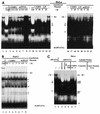
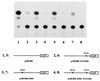
The human papillomavirus type 18 (HPV-18) upstream regulatory region (URR) controls cell type-specific expression of viral oncoproteins E6 and E7. The HPV-18 URR is highly active in HeLa cells, but its activity is virtually undetectable in HepG2 cells. Previous work has shown that YY1 plays an important role in activation of the HPV-18 URR in HeLa cells, and this activating activity is dependent on its physical interaction with C/EBPbeta, which binds to the switch region adjacent to the YY1 site in the URR. Overexpression of C/EBPbeta in HepG2 cells restores C/EBPbeta-YY1 interaction, resulting in strong activation of the HPV-18 URR activity. In this report, we show that, in contrast to the effect in HepG2 cells, overexpression of C/EBPbeta represses the HPV-18 URR in HeLa cells. This C/EBPbeta-induced repression of the HPV-18 URR in HeLa cells is binding site independent. It is also promoter specific, since it activates the albumin promoter under conditions in which it represses the URR in the same cells. Biochemical analysis shows that overexpression of C/EBPbeta in HeLa cells specifically interferes with binding of TATA-binding protein to the TATA box of the HPV-18 URR, but its overexpression in HepG2 cells leads to activation of the HPV-18 URR. These results suggest that a molecular mechanism underlies the ability of C/EBPbeta to regulate transcription in a cell type-specific manner and indicate the potential of using C/EBPbeta to manipulate the activity of the HPV-18 URR in cervical carcinoma cells.
Figures
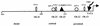



Similar articles
-
A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region.J Virol. 1996 Nov;70(11):7695-705. doi: 10.1128/JVI.70.11.7695-7705.1996. J Virol. 1996. PMID: 8892890 Free PMC article.
-
CCAAT/enhancer-binding protein beta represses human papillomavirus 11 upstream regulatory region expression through a promoter-proximal YY1-binding site.J Gen Virol. 2006 Jan;87(Pt 1):51-59. doi: 10.1099/vir.0.81207-0. J Gen Virol. 2006. PMID: 16361417
-
A switch region determines the cell type-specific positive or negative action of YY1 on the activity of the human papillomavirus type 18 promoter.J Virol. 1995 Jan;69(1):1-12. doi: 10.1128/JVI.69.1.1-12.1995. J Virol. 1995. PMID: 7983700 Free PMC article.
-
Silencing of integrated human papillomavirus type 18 oncogene transcription in cells expressing SerpinB2.J Virol. 2005 Apr;79(7):4246-56. doi: 10.1128/JVI.79.7.4246-4256.2005. J Virol. 2005. PMID: 15767426 Free PMC article.
-
Transcriptional control of human papillomavirus type 18 oncogene expression in different cell lines: role of transcription factor YY1.Virus Genes. 1995;11(1):53-8. doi: 10.1007/BF01701662. Virus Genes. 1995. PMID: 8808335
Cited by
-
Transcriptional regulation of mouse mast cell protease-2 by interleukin-15.J Biol Chem. 2009 Nov 20;284(47):32635-41. doi: 10.1074/jbc.M109.015446. Epub 2009 Oct 1. J Biol Chem. 2009. PMID: 19801677 Free PMC article.
-
Site-directed mutagenesis of human papillomavirus 18 promoter elements and tissue-specific expression in cervical carcinoma cells.Virus Genes. 2012 Jun;44(3):395-402. doi: 10.1007/s11262-012-0723-z. Epub 2012 Feb 21. Virus Genes. 2012. PMID: 22350992
-
Trichostatin A up-regulates human papillomavirus type 11 upstream regulatory region-E6 promoter activity in undifferentiated primary human keratinocytes.J Virol. 1999 Jun;73(6):5026-33. doi: 10.1128/JVI.73.6.5026-5033.1999. J Virol. 1999. PMID: 10233965 Free PMC article.
-
JAK/STAT Signaling and Cervical Cancer: From the Cell Surface to the Nucleus.Genes (Basel). 2023 May 24;14(6):1141. doi: 10.3390/genes14061141. Genes (Basel). 2023. PMID: 37372319 Free PMC article. Review.
-
Host cell restriction factors that limit transcription and replication of human papillomavirus.Virus Res. 2017 Mar 2;231:10-20. doi: 10.1016/j.virusres.2016.11.014. Epub 2016 Nov 15. Virus Res. 2017. PMID: 27863967 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

