A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly
- PMID: 9499062
- PMCID: PMC109501
- DOI: 10.1128/JVI.72.3.2072-2078.1998
A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly
Abstract
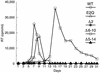
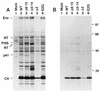
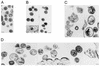
The capsid (CA) and nucleocapsid domains of the human immunodeficiency virus type 1 Gag polyprotein are separated by the p2 spacer peptide, which is essential for virus replication. Previous studies have revealed that p2 has an important role in virus morphogenesis. In this paper, we show that a crucial assembly determinant maps to the highly conserved N terminus of p2, which is predicted to form part of an alpha-helix that begins in CA. A mutational analysis indicates that the ability of the N terminus of p2 to adopt an alpha-helical structure is essential for its function during virus assembly. To prevent CA-p2 processing, it was necessary to mutate both the CA-p2 cleavage site and an internal cleavage site within p2. Virions produced by the double mutant lacked a conical core shell and instead contained a thin electron-dense shell about 10 nm underneath the virion membrane. These results suggest that p2 is transiently required for proper assembly, but needs to be removed from the C terminus of CA to weaken CA-CA interactions and allow the rearrangement of the virion core shell during virus maturation.
Figures

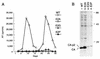

Similar articles
-
Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain.J Virol. 2000 Jun;74(12):5395-402. doi: 10.1128/jvi.74.12.5395-5402.2000. J Virol. 2000. PMID: 10823843 Free PMC article.
-
Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation.J Virol. 2006 Aug;80(16):7939-51. doi: 10.1128/JVI.00355-06. J Virol. 2006. PMID: 16873251 Free PMC article.
-
Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding.J Virol. 2000 Jun;74(11):5142-50. doi: 10.1128/jvi.74.11.5142-5150.2000. J Virol. 2000. PMID: 10799589 Free PMC article.
-
The Assembly of HTLV-1-How Does It Differ from HIV-1?Viruses. 2024 Sep 27;16(10):1528. doi: 10.3390/v16101528. Viruses. 2024. PMID: 39459862 Free PMC article. Review.
-
The choreography of HIV-1 proteolytic processing and virion assembly.J Biol Chem. 2012 Nov 30;287(49):40867-74. doi: 10.1074/jbc.R112.399444. Epub 2012 Oct 5. J Biol Chem. 2012. PMID: 23043111 Free PMC article. Review.
Cited by
-
Novel approaches to inhibiting HIV-1 replication.Antiviral Res. 2010 Jan;85(1):119-41. doi: 10.1016/j.antiviral.2009.09.009. Epub 2009 Sep 24. Antiviral Res. 2010. PMID: 19782103 Free PMC article. Review.
-
Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag.J Virol. 2005 Feb;79(3):1803-12. doi: 10.1128/JVI.79.3.1803-1812.2005. J Virol. 2005. PMID: 15650204 Free PMC article.
-
Inhibition of HIV Maturation via Selective Unfolding and Cross-Linking of Gag Polyprotein by a Mercaptobenzamide Acetylator.J Am Chem Soc. 2019 May 22;141(20):8327-8338. doi: 10.1021/jacs.9b02743. Epub 2019 May 13. J Am Chem Soc. 2019. PMID: 31042030 Free PMC article.
-
A single amino acid substitution in a segment of the CA protein within Gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome.J Virol. 2009 Jan;83(2):1105-14. doi: 10.1128/JVI.01439-08. Epub 2008 Nov 12. J Virol. 2009. PMID: 19004950 Free PMC article.
-
Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab.EMBO J. 1999 Mar 1;18(5):1124-36. doi: 10.1093/emboj/18.5.1124. EMBO J. 1999. PMID: 10064580 Free PMC article.
References
-
- Accola, M. A., and H. G. Göttlinger. Unpublished observation.
-
- Blaber M, Zhang X-J, Matthews B W. Structural basis of amino acid α helix propensity. Science. 1993;260:1637–1640. - PubMed
-
- Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Protein design: a hierarchic approach. Science. 1995;270:935–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

