Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry
- PMID: 9499048
- PMCID: PMC109487
- DOI: 10.1128/JVI.72.3.1949-1958.1998
Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry
Abstract
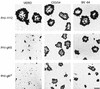
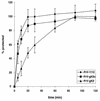
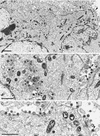
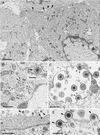
The pseudorabies virus (PrV) gene homologous to herpes simplex virus type 1 (HSV-1) UL53, which encodes HSV-1 glycoprotein K (gK), has recently been sequenced (J. Baumeister, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 69:5560-5567, 1995). To identify the corresponding protein, a rabbit antiserum was raised against a 40-kDa glutathione S-transferase-gK fusion protein expressed in Escherichia coli. In Western blot analysis, this serum detected a 32-kDa polypeptide in PrV-infected cell lysates as well as a 36-kDa protein in purified virion preparations, demonstrating that PrV gK is a structural component of virions. After treatment of purified virions with endoglycosidase H, a 34-kDa protein was detected, while after incubation with N-glycosidase F, a 32-kDa protein was specifically recognized. This finding indicates that virion gK is modified by N-linked glycans of complex as well as high-mannose type. For functional analysis, the UL53 open reading frame was interrupted after codon 164 by insertion of a gG-lacZ expression cassette into the wild-type PrV genome (PrV-gKbeta) or by insertion of the bovine herpesvirus 1 gB gene into a PrV gB- genome (PrV-gK(gB)). Infectious mutant virus progeny was obtained only on complementing gK-expressing cells, suggesting that gK has an important function in the replication cycle. After infection of Vero cells with either gK mutant, only single infected cells or small foci of infected cells were visible. In addition, virus yield was reduced approximately 30-fold, and penetration kinetics showed a delay in entry which could be compensated for by phenotypic gK complementation. Interestingly, the plating efficiency of PrV-gKbeta was similar to that of wild-type PrV on complementing and noncomplementing cells, pointing to an essential function of gK in virus egress but not entry. Ultrastructurally, virus assembly and morphogenesis of PrV gK mutants in noncomplementing cells were similar to wild-type virus. However, late in infection, numerous nucleocapsids were found directly underneath the plasma membrane in stages typical for the entry process, a phenomenon not observed after wild-type virus infection and also not visible after infection of gK-complementing cells. Thus, we postulate that presence of gK is important to inhibit immediate reinfection.
Figures



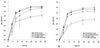
Similar articles
-
Pseudorabies virus glycoprotein K requires the UL20 gene product for processing.J Virol. 2000 Jun;74(11):5083-90. doi: 10.1128/jvi.74.11.5083-5090.2000. J Virol. 2000. PMID: 10799582 Free PMC article.
-
Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H.J Virol. 1997 Oct;71(10):7687-95. doi: 10.1128/JVI.71.10.7687-7695.1997. J Virol. 1997. PMID: 9311852 Free PMC article.
-
Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus.J Virol. 1999 Jul;73(7):5364-72. doi: 10.1128/JVI.73.7.5364-5372.1999. J Virol. 1999. PMID: 10364283 Free PMC article.
-
Host cellular factors involved in pseudorabies virus attachment and entry: a mini review.Front Vet Sci. 2023 Nov 27;10:1314624. doi: 10.3389/fvets.2023.1314624. eCollection 2023. Front Vet Sci. 2023. PMID: 38089700 Free PMC article. Review.
-
The Roles of Envelope Glycoprotein M in the Life Cycle of Some Alphaherpesviruses.Front Microbiol. 2021 Feb 19;12:631523. doi: 10.3389/fmicb.2021.631523. eCollection 2021. Front Microbiol. 2021. PMID: 33679658 Free PMC article. Review.
Cited by
-
Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization.J Virol. 2004 Dec;78(23):13262-77. doi: 10.1128/JVI.78.23.13262-13277.2004. J Virol. 2004. PMID: 15542677 Free PMC article.
-
The role of equine herpesvirus type 4 glycoprotein k in virus replication.Viruses. 2012 Aug;4(8):1258-63. doi: 10.3390/v4081258. Epub 2012 Aug 7. Viruses. 2012. PMID: 23012623 Free PMC article.
-
Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion.J Virol. 2003 Jan;77(1):499-510. doi: 10.1128/jvi.77.1.499-510.2003. J Virol. 2003. PMID: 12477855 Free PMC article.
-
Identification of non-essential loci within the Meleagrid herpesvirus 1 genome.Virol J. 2015 Aug 27;12:130. doi: 10.1186/s12985-015-0362-9. Virol J. 2015. PMID: 26307059 Free PMC article.
-
The egress of herpesviruses from cells: the unanswered questions.J Virol. 2006 Jul;80(13):6716-7; author replies 6717-9. doi: 10.1128/JVI.00386-06. J Virol. 2006. PMID: 16775362 Free PMC article. No abstract available.
References
-
- Baumeister, J., and B. G. Klupp. Unpublished data.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

