Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene
- PMID: 9454838
- PMCID: PMC6792720
- DOI: 10.1523/JNEUROSCI.18-04-01280.1998
Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene
Abstract
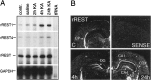
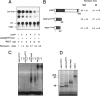
The identification of a common cis-acting silencer element, a neuron-restrictive silencer element (NRSE), in multiple neuron-specific genes, together with the finding that zinc finger transcription factor REST/NRSF/XBR could confer NRSE-mediated silencing in non-neuronal cells, suggested that REST/NRSF/XBR is a master negative regulator of neurogenesis. Here we show that, although REST/NRSF/XBR expression decreases during neuronal development, it proceeds in the adult nervous system. In situ hybridization analysis revealed neuronal expression of rat REST/NRSF/XBR mRNA in adult brain, with the highest levels in the neurons of hippocampus, pons/medulla, and midbrain. The glutamate analog kainic acid increased REST/NRSF/XBR mRNA levels in various hippocampal and cortical neurons in vivo, suggesting that REST/NRSF/XBR has a role in neuronal activity-implied processes. Several alternatively spliced REST/NRSF/XBR mRNAs encoding proteins with nine, five, or four zinc finger motifs are transcribed from REST/NRSF/XBR gene. Two of these transcripts are generated by neuron-specific splicing of a 28-bp-long exon. Rat REST/NRSF/XBR protein isoforms differ in their DNA binding specificities; however, all mediate repression in transient expression assays. Our data suggest that REST/NRSF/XBR is a negative regulator rather than a transcriptional silencer of neuronal gene expression and counteracts with positive regulators to modulate target gene expression quantitatively in different cell types, including neurons.
Figures




Similar articles
-
Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat.Brain Res Mol Brain Res. 1999 Sep 8;72(1):30-9. doi: 10.1016/s0169-328x(99)00196-5. Brain Res Mol Brain Res. 1999. PMID: 10521596
-
Expression of the rat m4 muscarinic acetylcholine receptor gene is regulated by the neuron-restrictive silencer element/repressor element 1.J Biol Chem. 1997 Feb 28;272(9):5854-60. doi: 10.1074/jbc.272.9.5854. J Biol Chem. 1997. PMID: 9038202
-
NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis.Nat Genet. 1998 Oct;20(2):136-42. doi: 10.1038/2431. Nat Genet. 1998. PMID: 9771705
-
Regulation of the cholinergic gene locus by the repressor element-1 silencing transcription factor/neuron restrictive silencer factor (REST/NRSF).Life Sci. 2004 Mar 19;74(18):2213-25. doi: 10.1016/j.lfs.2003.08.045. Life Sci. 2004. PMID: 15017977 Review.
-
Roles of the Neuron-Restrictive Silencer Factor in the Pathophysiological Process of the Central Nervous System.Front Cell Dev Biol. 2022 Mar 1;10:834620. doi: 10.3389/fcell.2022.834620. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35300407 Free PMC article. Review.
Cited by
-
G9a histone methyltransferase activity in retinal progenitors is essential for proper differentiation and survival of mouse retinal cells.J Neurosci. 2012 Dec 5;32(49):17658-70. doi: 10.1523/JNEUROSCI.1869-12.2012. J Neurosci. 2012. PMID: 23223288 Free PMC article.
-
Epigenetic mechanisms in neurological and neurodegenerative diseases.Front Cell Neurosci. 2015 Feb 27;9:58. doi: 10.3389/fncel.2015.00058. eCollection 2015. Front Cell Neurosci. 2015. PMID: 25774124 Free PMC article. Review.
-
Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression.Genome Biol. 2006;7(9):R85. doi: 10.1186/gb-2006-7-9-r85. Genome Biol. 2006. PMID: 17002790 Free PMC article.
-
Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation.Mol Cell Biol. 2004 Sep;24(18):8018-25. doi: 10.1128/MCB.24.18.8018-8025.2004. Mol Cell Biol. 2004. PMID: 15340064 Free PMC article.
-
MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression.J Biol Chem. 2009 Jan 30;284(5):2648-2656. doi: 10.1074/jbc.M806514200. Epub 2008 Dec 2. J Biol Chem. 2009. PMID: 19049968 Free PMC article.
References
-
- Abe H, Watanabe M, Yamakuni T, Kuwano R, Takahashi Y, Kondo H. Localization of gene expression of calbindin in the brain of adult rats. Neurosci Lett. 1992;138:211–215. - PubMed
-
- Barker PA, Lomen-Hoerth C, Gensch EM, Meakin S, Glass DJ, Shooter E. Tissue-specific alternative splicing generates two isoforms of the trkA receptor. J Biol Chem. 1993;268:15150–15157. - PubMed
-
- Belluardo N, Wu G, Mudo G, Hansson AC, Petterson R, Fuxe K. Comparative localization of fibroblast growth factor receptor -1, -2, and -3 mRNAs in the rat brain: in situ hybridization analysis. J Comp Neurol. 1997;379:226–246. - PubMed
-
- Ben-Ari Y, Represa A. Brief seizure episodes induce long-term potentiation and mossy fibre sprouting in the hippocampus. Trends Neurosci. 1990;13:375–403. - PubMed
-
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The proteins Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
