Tumorigenic potential of a recombinant retrovirus containing sequences from Moloney murine leukemia virus and feline leukemia virus
- PMID: 9445002
- PMCID: PMC124580
- DOI: 10.1128/JVI.72.2.1078-1084.1998
Tumorigenic potential of a recombinant retrovirus containing sequences from Moloney murine leukemia virus and feline leukemia virus
Erratum in
- J Virol. 2007 Jun;81(11):6158. Granger, S [corrected to Granger, S W]
Abstract
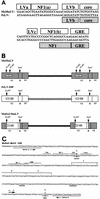
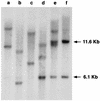
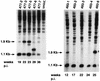
A recombinant retrovirus, termed MoFe2-MuLV, was constructed in which the U3 region of T-lymphomagenic Moloney murine leukemia virus (Mo-MuLV) was replaced by that of FeLV-945, a provirus of unique long terminal repeat (LTR) structure identified only in non-T-cell, non-B-cell lymphomas of the domestic cat. The LTR of FeLV-945 is unusual in that it contains only a single copy of the transcriptional enhancer followed 25 bp downstream by a 21-bp sequence in triplicate in tandem. Infectivity of MoFe2-MuLV was demonstrated in vitro in SC-1 cells and in vivo in neonatal NIH-Swiss mice. Tumors occurred in MoFe2-MuLV-infected animals following a latency period of 4 to 10 months (average, 6 months). The results of Southern blot analysis of the T-cell receptor beta locus demonstrated that all tumors were lymphomas of T-cell origin. MoFe2-MuLV LTRs were amplified by PCR from tumor DNA and were characterized by nucleotide sequence analysis. LTRs from the tumors that occurred with relatively shorter latency predominantly retained the original MoFe2-MuLV sequence intact and unaltered. Tumors that occurred with relatively longer latency contained LTRs that also retained the 21-bp sequence triplication characteristic of the original virus but had acquired various duplications of enhancer sequences. The repeated identification of enhancer duplications in late-appearing tumors suggests that the duplication affords a selective advantage, although apparently not in the efficient induction of T-cell lymphoma. Proto-oncogenes known to be targets of insertional mutagenesis in the majority of Mo-MuLV-induced tumors or in feline non-T-cell, non-B-cell lymphomas were shown not to be rearranged in any tumor examined. Mink cell focus-inducing (MCF) proviral DNA was readily detectable in some, but not all, tumors. The presence or absence of MCF did not correlate with the kinetics of tumor induction. These studies indicate that the single-enhancer, triplication-containing FeLV LTR, typical of non-T-cell, non-B-cell lymphomas in cats, is competent in the induction of T-cell lymphoma in mice. The findings suggest that the mechanism of MoFe2-MuLV-mediated lymphomagenesis may differ from that of Mo-MuLV-mediated disease, considering the possible involvement of novel oncogenes and the variable presence of MCF recombinants.
Figures
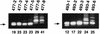

Similar articles
-
Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus.J Virol. 2002 Feb;76(3):1527-32. doi: 10.1128/jvi.76.3.1527-1532.2002. J Virol. 2002. PMID: 11773427 Free PMC article.
-
Substitution of feline leukemia virus long terminal repeat sequences into murine leukemia virus alters the pattern of insertional activation and identifies new common insertion sites.J Virol. 2005 Jan;79(1):57-66. doi: 10.1128/JVI.79.1.57-66.2005. J Virol. 2005. PMID: 15596801 Free PMC article.
-
Escape from in vivo restriction of Moloney mink cell focus-inducing viruses driven by the Mo+PyF101 long terminal repeat (LTR) by LTR alterations.J Virol. 1993 Dec;67(12):7140-8. doi: 10.1128/JVI.67.12.7140-7148.1993. J Virol. 1993. PMID: 8230436 Free PMC article.
-
Advances in understanding molecular determinants in FeLV pathology.Vet Immunol Immunopathol. 2008 May 15;123(1-2):14-22. doi: 10.1016/j.vetimm.2008.01.008. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18289704 Free PMC article. Review.
-
Endogenous env elements: partners in generation of pathogenic feline leukemia viruses.Virus Genes. 1995;11(2-3):147-61. doi: 10.1007/BF01728655. Virus Genes. 1995. PMID: 8828142 Review.
Cited by
-
Matrix attachment regions as targets for retroviral integration.Virol J. 2005 Aug 19;2:68. doi: 10.1186/1743-422X-2-68. Virol J. 2005. PMID: 16111492 Free PMC article.
-
Feline leukemia virus long terminal repeat activates collagenase IV gene expression through AP-1.J Virol. 1999 Jun;73(6):4931-40. doi: 10.1128/JVI.73.6.4931-4940.1999. J Virol. 1999. PMID: 10233955 Free PMC article.
-
Tandemization of a subregion of the enhancer sequences from SRS 19-6 murine leukemia virus associated with T-lymphoid but not other leukemias.J Virol. 1999 Sep;73(9):7175-84. doi: 10.1128/JVI.73.9.7175-7184.1999. J Virol. 1999. PMID: 10438804 Free PMC article.
-
Viral determinants of FeLV infection and pathogenesis: lessons learned from analysis of a natural cohort.Viruses. 2011 Sep;3(9):1681-98. doi: 10.3390/v3091681. Epub 2011 Sep 9. Viruses. 2011. PMID: 21994802 Free PMC article. Review.
-
Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus.J Virol. 2002 Feb;76(3):1527-32. doi: 10.1128/jvi.76.3.1527-1532.2002. J Virol. 2002. PMID: 11773427 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

