Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells
- PMID: 9420220
- PMCID: PMC109369
- DOI: 10.1128/JVI.72.1.236-244.1998
Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells
Abstract
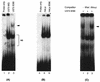
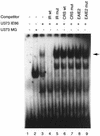
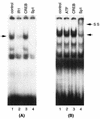
Human cytomegalovirus (HCMV) gene expression is highly cell and tissue specific. Cell factor-mediated regulatory interactions are involved in regulating the restricted expression of the HCMV major immediate-early (IE) gene (J. F. Baskar, P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson, 70:3207-3213, 1996). To gain an understanding of HCMV early gene activation, we studied the effect of each of the three major IE proteins, IE72, IE86, and IE55, on the HCMV DNA polymerase gene (pol; UL54) promoter. In transient-expression assays, the IE86 protein alone was able to transactivate the pol promoter, but IE72 and IE55 were not, in permissive U373MG cells. However, we were unable to detect IE86-mediated transactivation in nonpermissive HeLa or C33-A cells. Using electrophoretic mobility shift assays (EMSAs), we found that expression of the IE86 protein in U373MG cells resulted in specific binding of a DNA complex to an inverted-repeat element, IR1, of the pol promoter. Antibody supershifting and EMSA-Western blotting experiments further showed that IE86 and the cellular transcription factor Sp1 were components of the IR1 DNA-binding complex. Furthermore, we found that binding of DNA by Sp1 was dramatically increased in the presence of IE86. Interestingly, this IE86-induced DNA-binding activity of Sp1 was inhibited by a repressor activity presented in HeLa cells. In summary, our study suggests that a viral regulatory protein can modulate the DNA binding activity of a cellular transcription factor, resulting in cell-specific transactivation of viral genes.
Figures





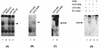

Similar articles
-
Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter.J Virol. 1997 Sep;71(9):6683-91. doi: 10.1128/JVI.71.9.6683-6691.1997. J Virol. 1997. PMID: 9261391 Free PMC article.
-
Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB.J Virol. 1995 Oct;69(10):6030-7. doi: 10.1128/JVI.69.10.6030-6037.1995. J Virol. 1995. PMID: 7666507 Free PMC article.
-
The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells.J Virol. 2000 Aug;74(15):7108-18. doi: 10.1128/jvi.74.15.7108-7118.2000. J Virol. 2000. PMID: 10888651 Free PMC article.
-
Activation and regulation of human cytomegalovirus early genes.Intervirology. 1996;39(5-6):361-77. doi: 10.1159/000150507. Intervirology. 1996. PMID: 9130046 Review.
-
Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate.Clin Microbiol Rev. 1999 Jul;12(3):367-82. doi: 10.1128/CMR.12.3.367. Clin Microbiol Rev. 1999. PMID: 10398670 Free PMC article. Review.
Cited by
-
Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes.J Virol. 2001 Feb;75(4):1870-8. doi: 10.1128/JVI.75.4.1870-1878.2001. J Virol. 2001. PMID: 11160686 Free PMC article.
-
Activation of transcription of the human cytomegalovirus early UL4 promoter by the Ets transcription factor binding element.J Virol. 2000 Nov;74(21):9845-57. doi: 10.1128/jvi.74.21.9845-9857.2000. J Virol. 2000. PMID: 11024111 Free PMC article.
-
NF-kappaB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86.J Virol. 2008 May;82(9):4250-6. doi: 10.1128/JVI.02156-07. Epub 2008 Feb 20. J Virol. 2008. PMID: 18287226 Free PMC article.
-
The roles of viruses in brain tumor initiation and oncomodulation.J Neurooncol. 2011 Dec;105(3):451-66. doi: 10.1007/s11060-011-0658-6. Epub 2011 Jul 1. J Neurooncol. 2011. PMID: 21720806 Free PMC article. Review.
-
The 6-aminoquinolone WC5 inhibits human cytomegalovirus replication at an early stage by interfering with the transactivating activity of viral immediate-early 2 protein.Antimicrob Agents Chemother. 2010 May;54(5):1930-40. doi: 10.1128/AAC.01730-09. Epub 2010 Mar 1. Antimicrob Agents Chemother. 2010. PMID: 20194695 Free PMC article.
References
-
- Baskar J F, Smith P P, Nilaver G, Jupp R A, Hoffmann S, Peffer N J, Tenney D J, Colberg-Poley A M, Ghazal P, Nelson J A. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. - PMC - PubMed
-
- Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. - PubMed
-
- Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor. Science. 1986;234:47–52. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

