Regulation of sex-specific selection of fruitless 5' splice sites by transformer and transformer-2
- PMID: 9418892
- PMCID: PMC121514
- DOI: 10.1128/MCB.18.1.450
Regulation of sex-specific selection of fruitless 5' splice sites by transformer and transformer-2
Abstract
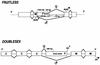
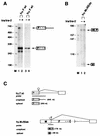
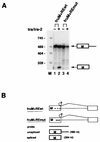
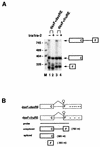
In Drosophila melanogaster, the fruitless (fru) gene controls essentially all aspects of male courtship behavior. It does this through sex-specific alternative splicing of the fru pre-mRNA, leading to the production of male-specific fru mRNAs capable of expressing male-specific fru proteins. Sex-specific fru splicing involves the choice between alternative 5' splice sites, one used exclusively in males and the other used only in females. Here we report that the Drosophila sex determination genes transformer (tra) and transformer-2 (tra-2) switch fru splicing from the male-specific pattern to the female-specific pattern through activation of the female-specific fru 5' splice site. Activation of female-specific fru splicing requires cis-acting tra and tra-2 repeat elements that are part of an exonic splicing enhancer located immediately upstream of the female-specific fru 5' splice site and are recognized by the TRA and TRA-2 proteins in vitro. This fru splicing enhancer is sufficient to promote the activation by tra and tra-2 of both a 5' splice site and the female-specific doublesex (dsx) 3' splice site, suggesting that the mechanisms of 5' splice site activation and 3' splice site activation may be similar.
Figures


Similar articles
-
Enhancer-dependent 5'-splice site control of fruitless pre-mRNA splicing.J Biol Chem. 2003 Jun 20;278(25):22740-7. doi: 10.1074/jbc.M301036200. Epub 2003 Mar 19. J Biol Chem. 2003. PMID: 12646561 Free PMC article.
-
The transformer-2 and fruitless characterisation with developmental expression profiles of sex-determining genes in Bactrocera dorsalis and B. correcta.Sci Rep. 2020 Oct 21;10(1):17938. doi: 10.1038/s41598-020-74856-6. Sci Rep. 2020. PMID: 33087807 Free PMC article.
-
Tribolium castaneum Transformer-2 regulates sex determination and development in both males and females.Insect Biochem Mol Biol. 2013 Dec;43(12):1125-32. doi: 10.1016/j.ibmb.2013.08.010. Epub 2013 Sep 19. Insect Biochem Mol Biol. 2013. PMID: 24056158 Free PMC article.
-
Fruitless alternative splicing and sex behaviour in insects: an ancient and unforgettable love story?J Genet. 2010 Sep;89(3):287-99. doi: 10.1007/s12041-010-0040-z. J Genet. 2010. PMID: 20876995 Review.
-
From behavior to development: genes for sexual behavior define the neuronal sexual switch in Drosophila.Mech Dev. 1998 May;73(2):135-46. doi: 10.1016/s0925-4773(98)00042-2. Mech Dev. 1998. PMID: 9622612 Review.
Cited by
-
Systems behavior: of male courtship, the nervous system and beyond in Drosophila.Curr Genomics. 2008 Dec;9(8):517-24. doi: 10.2174/138920208786847980. Curr Genomics. 2008. PMID: 19516958 Free PMC article.
-
Neuroethology of male courtship in Drosophila: from the gene to behavior.J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014 Apr;200(4):251-64. doi: 10.1007/s00359-014-0891-5. Epub 2014 Feb 25. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014. PMID: 24567257 Review.
-
Tra2beta as a novel mediator of vascular smooth muscle diversification.Circ Res. 2008 Aug 29;103(5):485-92. doi: 10.1161/CIRCRESAHA.108.178384. Epub 2008 Jul 31. Circ Res. 2008. PMID: 18669920 Free PMC article.
-
Functional dissection of the neural substrates for sexual behaviors in Drosophila melanogaster.Genetics. 2011 Sep;189(1):195-211. doi: 10.1534/genetics.111.129940. Epub 2011 Jul 29. Genetics. 2011. PMID: 21705753 Free PMC article.
-
Alcohol-induced aggression in Drosophila.Addict Biol. 2021 Sep;26(5):e13045. doi: 10.1111/adb.13045. Epub 2021 May 27. Addict Biol. 2021. PMID: 34044470 Free PMC article.
References
-
- Aebi M, Hornig H, Weissman C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. - PubMed
-
- Amrein H, Gorman M, Nöthiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. - PubMed
-
- Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. - PubMed
-
- Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
