Histone acetyltransferases regulate HIV-1 enhancer activity in vitro
- PMID: 9407026
- PMCID: PMC316802
- DOI: 10.1101/gad.11.24.3327
Histone acetyltransferases regulate HIV-1 enhancer activity in vitro
Abstract
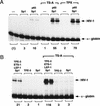
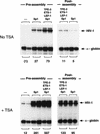
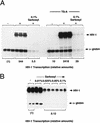
Specific inhibitors of histone deacetylase, such as trichostatin A (TSA) and trapoxin (TPX), are potent inducers of HIV-1 transcription in latently infected T-cell lines. Activation of the integrated HIV-1 promoter is accompanied by the loss or rearrangement of a positioned nucleosome (nuc-1) near the viral RNA start site. Here we show that TSA strongly induces HIV-1 transcription on chromatin in vitro, concomitant with an enhancer factor-assisted increase in the level of acetylated histone H4. TSA treatment, however, did not detectably alter enhancer factor binding or the positioning of nuc-1 on the majority of the chromatin templates indicating that protein acetylation and chromatin remodeling may be limiting steps that occur only on transcriptionally competent templates, or that remodeling of nuc-1 requires additional factors. To assess the number of active chromatin templates in vitro, transcription was limited to a single round with low levels of the detergent Sarkosyl. Remarkably, HIV-1 transcription on chromatin was found to arise from a small number of active templates that can each support nearly 100 rounds of transcription, and TSA increased the number of active templates in each round. In contrast, transcription on naked DNA was limited to only a few rounds and was not responsive to TSA. We conclude that HIV-1 enhancer complexes greatly facilitate transcription reinitiation on chromatin in vitro, and act at a limiting step to promote the acetylation of histones or other transcription factors required for HIV-1 enhancer activity.
Figures





Similar articles
-
Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation.EMBO J. 1996 Mar 1;15(5):1112-20. EMBO J. 1996. PMID: 8605881 Free PMC article.
-
Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat.Mol Cell Biol. 2002 May;22(9):2965-73. doi: 10.1128/MCB.22.9.2965-2973.2002. Mol Cell Biol. 2002. PMID: 11940654 Free PMC article.
-
Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure.Mol Cell Biol. 2002 Dec;22(23):8302-19. doi: 10.1128/MCB.22.23.8302-8319.2002. Mol Cell Biol. 2002. PMID: 12417732 Free PMC article.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
-
Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin.Mol Biol Rep. 1997 Aug;24(3):197-207. doi: 10.1023/a:1006811817247. Mol Biol Rep. 1997. PMID: 9291093 Review.
Cited by
-
ORC1 enhances repressive epigenetic modifications on HIV-1 LTR to promote HIV-1 latency.J Virol. 2024 Aug 20;98(8):e0003524. doi: 10.1128/jvi.00035-24. Epub 2024 Jul 31. J Virol. 2024. PMID: 39082875
-
Novel assays to investigate the mechanisms of latent infection with HIV-2.PLoS One. 2022 Apr 27;17(4):e0267402. doi: 10.1371/journal.pone.0267402. eCollection 2022. PLoS One. 2022. PMID: 35476802 Free PMC article.
-
Lentiviral Nef Proteins Differentially Govern the Establishment of Viral Latency.J Virol. 2022 Apr 13;96(7):e0220621. doi: 10.1128/jvi.02206-21. Epub 2022 Mar 10. J Virol. 2022. PMID: 35266804 Free PMC article.
-
Host T Cell Dedifferentiation Effects Drive HIV-1 Latency Stability.J Virol. 2022 Mar 9;96(5):e0197421. doi: 10.1128/jvi.01974-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019721 Free PMC article.
-
Balance between Retroviral Latency and Transcription: Based on HIV Model.Pathogens. 2020 Dec 29;10(1):16. doi: 10.3390/pathogens10010016. Pathogens. 2020. PMID: 33383617 Free PMC article. Review.
References
-
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;89:341–347. - PubMed
-
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmonson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. - PubMed
-
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
