Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex
- PMID: 9245791
- PMCID: PMC2141630
- DOI: 10.1083/jcb.138.3.629
Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex
Abstract
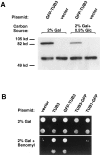
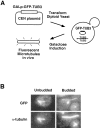
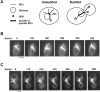
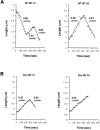
Proper orientation of the mitotic spindle is critical for successful cell division in budding yeast. To investigate the mechanism of spindle orientation, we used a green fluorescent protein (GFP)-tubulin fusion protein to observe microtubules in living yeast cells. GFP-tubulin is incorporated into microtubules, allowing visualization of both cytoplasmic and spindle microtubules, and does not interfere with normal microtubule function. Microtubules in yeast cells exhibit dynamic instability, although they grow and shrink more slowly than microtubules in animal cells. The dynamic properties of yeast microtubules are modulated during the cell cycle. The behavior of cytoplasmic microtubules revealed distinct interactions with the cell cortex that result in associated spindle movement and orientation. Dynein-mutant cells had defects in these cortical interactions, resulting in misoriented spindles. In addition, microtubule dynamics were altered in the absence of dynein. These results indicate that microtubules and dynein interact to produce dynamic cortical interactions, and that these interactions result in the force driving spindle orientation.
Figures

Similar articles
-
Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud.J Cell Biol. 1997 Nov 17;139(4):985-94. doi: 10.1083/jcb.139.4.985. J Cell Biol. 1997. PMID: 9362516 Free PMC article.
-
The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast.J Cell Biol. 2003 Feb 3;160(3):355-64. doi: 10.1083/jcb.200209022. J Cell Biol. 2003. PMID: 12566428 Free PMC article.
-
Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae.J Cell Biol. 1995 Aug;130(3):687-700. doi: 10.1083/jcb.130.3.687. J Cell Biol. 1995. PMID: 7622568 Free PMC article.
-
Using green fluorescent protein fusion proteins to quantitate microtubule and spindle dynamics in budding yeast.Methods Cell Biol. 1999;61:369-83. doi: 10.1016/s0091-679x(08)61990-1. Methods Cell Biol. 1999. PMID: 9891324 Review. No abstract available.
-
Mechanisms of spindle positioning.J Cell Biol. 2013 Jan 21;200(2):131-40. doi: 10.1083/jcb.201210007. J Cell Biol. 2013. PMID: 23337115 Free PMC article. Review.
Cited by
-
Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation.Cell. 2012 Mar 2;148(5):958-72. doi: 10.1016/j.cell.2012.01.041. Cell. 2012. PMID: 22385961 Free PMC article.
-
Reconstitution of Basic Mitotic Spindles in Spherical Emulsion Droplets.J Vis Exp. 2016 Aug 13;(114):54278. doi: 10.3791/54278. J Vis Exp. 2016. PMID: 27584979 Free PMC article.
-
SCAR/WAVE is activated at mitosis and drives myosin-independent cytokinesis.J Cell Sci. 2010 Jul 1;123(Pt 13):2246-55. doi: 10.1242/jcs.063735. Epub 2010 Jun 8. J Cell Sci. 2010. PMID: 20530573 Free PMC article.
-
Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque.Mol Biol Cell. 2001 Aug;12(8):2519-33. doi: 10.1091/mbc.12.8.2519. Mol Biol Cell. 2001. PMID: 11514632 Free PMC article.
-
beta-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast.Mol Biol Cell. 2002 Aug;13(8):2919-32. doi: 10.1091/mbc.e02-01-0003. Mol Biol Cell. 2002. PMID: 12181356 Free PMC article.
References
-
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. - PubMed
-
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

