Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90
- PMID: 8962087
- PMCID: PMC26168
- DOI: 10.1073/pnas.93.25.14536
Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90
Abstract
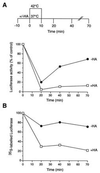
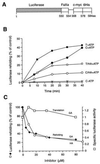
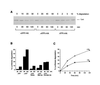
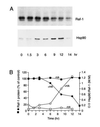
The role of the abundant stress protein Hsp90 in protecting cells against stress-induced damage is not well understood. The recent discovery that a class of ansamycin antibiotics bind specifically to Hsp90 allowed us to address this problem from a new angle. We find that mammalian Hsp90, in cooperation with Hsp70, p60, and other factors, mediates the ATP-dependent refolding of heat-denatured proteins, such as firefly luciferase. Failure to refold results in proteolysis. The ansamycins inhibit refolding, both in vivo and in a cell extract, by preventing normal dissociation of Hsp90 from luciferase, causing its enhanced degradation. This mechanism also explains the ansamycin-induced proteolysis of several protooncogenic protein kinases, such as Raf-1, which interact with Hsp90. We propose that Hsp90 is part of a quality control system that facilitates protein refolding or degradation during recovery from stress. This function is used by a limited set of signal transduction molecules for their folding and regulation under nonstress conditions. The ansamycins shift the mode of Hsp90 from refolding to degradation, and this effect is probably amplified for specific Hsp90 substrates.
Figures


Similar articles
-
Hsc70/Hsp40 chaperone system mediates the Hsp90-dependent refolding of firefly luciferase.Genes Cells. 1999 Dec;4(12):721-9. doi: 10.1046/j.1365-2443.1999.00299.x. Genes Cells. 1999. PMID: 10620017
-
A critical role for the proteasome activator PA28 in the Hsp90-dependent protein refolding.J Biol Chem. 2000 Mar 24;275(12):9055-61. doi: 10.1074/jbc.275.12.9055. J Biol Chem. 2000. PMID: 10722756
-
Effect of geldanamycin on the kinetics of chaperone-mediated renaturation of firefly luciferase in rabbit reticulocyte lysate.Biochemistry. 1996 Oct 15;35(41):13443-50. doi: 10.1021/bi9615396. Biochemistry. 1996. PMID: 8873613
-
Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation.Med Res Rev. 2006 May;26(3):310-38. doi: 10.1002/med.20052. Med Res Rev. 2006. PMID: 16385472 Review.
-
Hsp90: a chaperone for protein folding and gene regulation.Biochem Cell Biol. 2005 Dec;83(6):703-10. doi: 10.1139/o05-158. Biochem Cell Biol. 2005. PMID: 16333321 Review.
Cited by
-
An Hsp90 modulator that exhibits a unique mechanistic profile.Bioorg Med Chem Lett. 2012 May 1;22(9):3287-90. doi: 10.1016/j.bmcl.2012.03.012. Epub 2012 Mar 11. Bioorg Med Chem Lett. 2012. PMID: 22480433 Free PMC article.
-
Induced protein degradation for therapeutics: past, present, and future.J Clin Invest. 2024 Jan 2;134(1):e175265. doi: 10.1172/JCI175265. J Clin Invest. 2024. PMID: 38165043 Free PMC article. Review.
-
ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system.Nat Chem Biol. 2013 May;9(5):307-12. doi: 10.1038/nchembio.1212. Epub 2013 Mar 17. Nat Chem Biol. 2013. PMID: 23502424 Free PMC article.
-
Heat shock protein 90 inhibition depletes TrkA levels and signaling in human acute leukemia cells.Mol Cancer Ther. 2010 Aug;9(8):2232-42. doi: 10.1158/1535-7163.MCT-10-0336. Epub 2010 Jul 27. Mol Cancer Ther. 2010. PMID: 20663926 Free PMC article.
-
Covalent inhibitors of EGFR family protein kinases induce degradation of human Tribbles 2 (TRIB2) pseudokinase in cancer cells.Sci Signal. 2018 Sep 25;11(549):eaat7951. doi: 10.1126/scisignal.aat7951. Sci Signal. 2018. PMID: 30254057 Free PMC article.
References
-
- Craig E A, Gross C A. Trends Biochem Sci. 1991;16:135–140. - PubMed
-
- Parsell D A, Lindquist S. Annu Rev Genet. 1993;27:437–496. - PubMed
-
- Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the Heatshock Proteins of Escherichia coli and the Autoregulation of the Heat Shock Response. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 209–249.
-
- Wiech H, Buchner J, Zimmermann R, Jacob U. Nature (London) 1992;358:169–170. - PubMed
-
- Yonehara M, Minami Y, Kawata Y, Nagai J, Yahara I. J Biol Chem. 1996;271:2641–2645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

