Partitioning of fatty acids between membrane and storage lipids controls ER membrane expansion
- PMID: 39753951
- PMCID: PMC11790888
- DOI: 10.1038/s44318-024-00355-3
Partitioning of fatty acids between membrane and storage lipids controls ER membrane expansion
Abstract
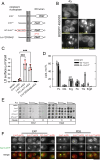
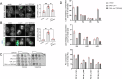
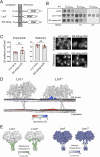
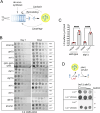
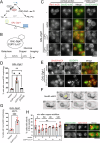
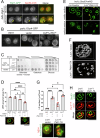
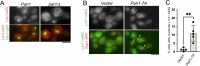
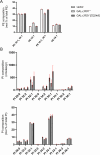
Biogenesis of membrane-bound organelles involves the synthesis, remodeling, and degradation of their constituent phospholipids. How these pathways regulate organelle size remains poorly understood. Here we demonstrate that a lipid-degradation pathway inhibits expansion of the endoplasmic reticulum (ER) membrane. Phospholipid diacylglycerol acyltransferases (PDATs) use endogenous phospholipids as fatty-acyl donors to generate triglyceride stored in lipid droplets. The significance of this non-canonical triglyceride biosynthesis pathway has remained elusive. We find that the activity of the yeast PDAT Lro1 is regulated by a membrane-proximal helical segment facing the luminal side of the ER bilayer. To reveal the biological roles of PDATs, we engineered an Lro1 variant with derepressed activity. We show that active Lro1 mediates retraction of ER membrane expansion driven by phospholipid synthesis. Furthermore, subcellular distribution and membrane turnover activity of Lro1 are controlled by diacylglycerol produced by the activity of Pah1, a conserved member of the lipin family. Collectively, our findings reveal a lipid-metabolic network that regulates endoplasmic reticulum biogenesis by converting phospholipids into storage lipids.
Keywords: Endoplasmic Reticulum; Lipid Droplet; Membrane; Phospholipid; Yeast.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures

Update of
-
The partitioning of fatty acids between membrane and storage lipids controls ER membrane expansion.bioRxiv [Preprint]. 2024 Sep 5:2024.09.05.611378. doi: 10.1101/2024.09.05.611378. bioRxiv. 2024. Update in: EMBO J. 2025 Feb;44(3):781-800. doi: 10.1038/s44318-024-00355-3. PMID: 39282465 Free PMC article. Updated. Preprint.
Similar articles
-
The partitioning of fatty acids between membrane and storage lipids controls ER membrane expansion.bioRxiv [Preprint]. 2024 Sep 5:2024.09.05.611378. doi: 10.1101/2024.09.05.611378. bioRxiv. 2024. Update in: EMBO J. 2025 Feb;44(3):781-800. doi: 10.1038/s44318-024-00355-3. PMID: 39282465 Free PMC article. Updated. Preprint.
-
Compartmentalized Synthesis of Triacylglycerol at the Inner Nuclear Membrane Regulates Nuclear Organization.Dev Cell. 2019 Sep 23;50(6):755-766.e6. doi: 10.1016/j.devcel.2019.07.009. Epub 2019 Aug 15. Dev Cell. 2019. PMID: 31422915 Free PMC article.
-
The Saccharomyces cerevisiae Spo7 basic tail is required for Nem1-Spo7/Pah1 phosphatase cascade function in lipid synthesis.J Biol Chem. 2024 Jan;300(1):105587. doi: 10.1016/j.jbc.2023.105587. Epub 2023 Dec 21. J Biol Chem. 2024. PMID: 38141768 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Mechanism of Obesity-Related Lipotoxicity and Clinical Perspective.Adv Exp Med Biol. 2024;1460:131-166. doi: 10.1007/978-3-031-63657-8_5. Adv Exp Med Biol. 2024. PMID: 39287851 Review.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
-
- Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A (2007) SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem 282:30845–30855 - PubMed
-
- Bernetti M, Bussi G (2020) Pressure control using stochastic cell rescaling. J Chem Phys 153:114107 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

