Localized molecular chaperone synthesis maintains neuronal dendrite proteostasis
- PMID: 39737952
- PMCID: PMC11685665
- DOI: 10.1038/s41467-024-55055-7
Localized molecular chaperone synthesis maintains neuronal dendrite proteostasis
Abstract
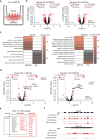
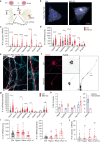
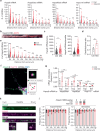
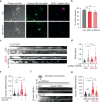
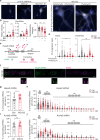
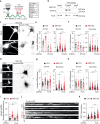
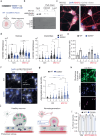
Proteostasis is maintained through regulated protein synthesis and degradation and chaperone-assisted protein folding. However, this is challenging in neuronal projections because of their polarized morphology and constant synaptic proteome remodeling. Using high-resolution fluorescence microscopy, we discover that hippocampal and spinal cord motor neurons of mouse and human origin localize a subset of chaperone mRNAs to their dendrites and use microtubule-based transport to increase this asymmetric localization following proteotoxic stress. The most abundant dendritic chaperone mRNA encodes a constitutive heat shock protein 70 family member (HSPA8). Proteotoxic stress also enhances HSPA8 mRNA translation efficiency in dendrites. Stress-mediated HSPA8 mRNA localization to the dendrites is impaired by depleting fused in sarcoma-an amyotrophic lateral sclerosis-related protein-in cultured spinal cord mouse motor neurons or by expressing a pathogenic variant of heterogenous nuclear ribonucleoprotein A2/B1 in neurons derived from human induced pluripotent stem cells. These results reveal a neuronal stress response in which RNA-binding proteins increase the dendritic localization of HSPA8 mRNA to maintain proteostasis and prevent neurodegeneration.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: G.W.Y. is a Scientific Advisory Board member of Jumpcode Genomics and a co-founder, Board of Directors, and Scientific Advisory Board member, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. The remaining authors declare no competing interests.
Figures
Update of
-
Localized synthesis of molecular chaperones sustains neuronal proteostasis.bioRxiv [Preprint]. 2024 Oct 19:2023.10.03.560761. doi: 10.1101/2023.10.03.560761. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10796. doi: 10.1038/s41467-024-55055-7. PMID: 37873158 Free PMC article. Updated. Preprint.
-
Localized molecular chaperone synthesis maintains neuronal dendrite proteostasis.Res Sq [Preprint]. 2023 Dec 11:rs.3.rs-3673702. doi: 10.21203/rs.3.rs-3673702/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Dec 30;15(1):10796. doi: 10.1038/s41467-024-55055-7. PMID: 38168440 Free PMC article. Updated. Preprint.
Similar articles
-
Localized molecular chaperone synthesis maintains neuronal dendrite proteostasis.Res Sq [Preprint]. 2023 Dec 11:rs.3.rs-3673702. doi: 10.21203/rs.3.rs-3673702/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Dec 30;15(1):10796. doi: 10.1038/s41467-024-55055-7. PMID: 38168440 Free PMC article. Updated. Preprint.
-
Localized synthesis of molecular chaperones sustains neuronal proteostasis.bioRxiv [Preprint]. 2024 Oct 19:2023.10.03.560761. doi: 10.1101/2023.10.03.560761. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10796. doi: 10.1038/s41467-024-55055-7. PMID: 37873158 Free PMC article. Updated. Preprint.
-
A variant of the Hspa8 synaptic chaperone modifies disease in a SOD1G86R mouse model of amyotrophic lateral sclerosis.Exp Neurol. 2025 Jan;383:115024. doi: 10.1016/j.expneurol.2024.115024. Epub 2024 Oct 24. Exp Neurol. 2025. PMID: 39454934
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
References
-
- Huber, K. M., Kayser, M. S. & Bear, M. F. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science288, 1254–1256 (2000). - PubMed
-
- Kang, H. & Schuman, E. M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science273, 1402–1406 (1996). - PubMed
-
- Miller, S. et al. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron36, 507–519 (2002). - PubMed
MeSH terms
Substances
Grants and funding
- RF1 MH126719/MH/NIMH NIH HHS/United States
- 300232/Fonds de Recherche du Québec - Santé (Fonds de la recherche en sante du Quebec)
- MH126719/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- HG009889/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U41 HG009889/HG/NHGRI NIH HHS/United States
- U24 HG009889/HG/NHGRI NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- HG011864/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- PJT-186141/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- HG004659/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- Hudson Translational Team Grant/ALS Society of Canada (ALS Canada)
- R01 HG011864/HG/NHGRI NIH HHS/United States
- R01 NS103172/NS/NINDS NIH HHS/United States
- 2022-ALS Discovery Grant/ALS Society of Canada (ALS Canada)
LinkOut - more resources
Full Text Sources
Miscellaneous

