Oral fungal dysbiosis and systemic immune dysfunction in Chinese patients with schizophrenia
- PMID: 39572530
- PMCID: PMC11582559
- DOI: 10.1038/s41398-024-03183-5
Oral fungal dysbiosis and systemic immune dysfunction in Chinese patients with schizophrenia
Abstract
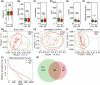
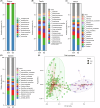
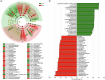
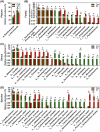
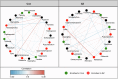
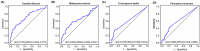
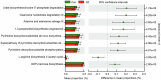
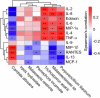
Oral microbial dysbiosis contributes to the development of schizophrenia (SZ). While numerous studies have investigated alterations in the oral bacterial microbiota among SZ patients, investigations into the fungal microbiota, another integral component of the oral microbiota, are scarce. In this cross-sectional study, we enrolled 118 Chinese patients with SZ and 97 age-matched healthy controls (HCs) to evaluate the oral fungal microbiota from tongue coating samples using internal transcribed spacer 1 amplicon sequencing and assess host immunity via multiplex immunoassays. Our findings revealed that SZ patients exhibited reduced fungal richness and significant differences in β-diversity compared to HCs. Within the oral fungal communities, we identified two distinct fungal clusters (mycotypes): Candida and Malassezia, with SZ patients showing increased Malassezia and decreased Candida levels. These key functional oral fungi may serve as potential diagnostic biomarkers for SZ. Furthermore, SZ patients displayed signs of immunological dysfunction, characterized by elevated levels of pro-inflammatory cytokines such as IL-6 and TNF-α, and chemokines including MIP-1α and MCP-1. Importantly, Malassezia mycotype correlated positively with peripheral pro-inflammatory cytokines, while Candida mycotype exhibited a negative correlation with these cytokines. In conclusion, we have demonstrated, for the first time, the presence of altered oral fungal communities and systemic immune dysfunction in Chinese SZ patients compared to HCs, providing novel insights into the potential role of oral fungi as biomarkers and the broader implications for understanding SZ pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Age-related differences in the interplay of fluency and complexity in Chinese-speaking seniors' oral narratives.Int J Lang Commun Disord. 2024 Sep-Oct;59(5):1672-1690. doi: 10.1111/1460-6984.13023. Epub 2024 Feb 26. Int J Lang Commun Disord. 2024. PMID: 38406919
-
Association between the oral microbiome and brain resting state connectivity in schizophrenia.Schizophr Res. 2024 Aug;270:392-402. doi: 10.1016/j.schres.2024.06.045. Epub 2024 Jul 9. Schizophr Res. 2024. PMID: 38986386
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Acupuncture for acute hordeolum.Cochrane Database Syst Rev. 2017 Feb 9;2(2):CD011075. doi: 10.1002/14651858.CD011075.pub2. Cochrane Database Syst Rev. 2017. PMID: 28181687 Free PMC article. Review.
References
-
- Solmi M, Seitidis G, Mavridis D, Correll CU, Dragioti E, Guimond S, et al. Incidence, prevalence, and global burden of schizophrenia - data, with critical appraisal, from the Global Burden of Disease (GBD) 2019. Mol Psychiatry. 2023;28:5319–27. - PubMed
-
- Marder SR, Cannon TD. Schizophrenia. N Engl J Med. 2019;381:1753–61. - PubMed
-
- Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, et al. Heritability of schizophrenia and schizophrenia spectrum based on the nationwide danish twin register. Biol Psychiatry. 2018;83:492–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

