Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes
- PMID: 39571015
- PMCID: PMC11583848
- DOI: 10.1126/science.adq8587
Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes
Abstract
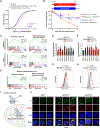
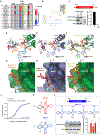
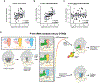
The CCR4-NOT complex is a major regulator of eukaryotic messenger RNA (mRNA) stability. Slow decoding during translation promotes association of CCR4-NOT with ribosomes, accelerating mRNA degradation. We applied selective ribosome profiling to further investigate the determinants of CCR4-NOT recruitment to ribosomes in mammalian cells. This revealed that specific arginine codons in the P-site are strong signals for ribosomal recruitment of human CNOT3, a CCR4-NOT subunit. Cryo-electron microscopy and transfer RNA (tRNA) mutagenesis demonstrated that the D-arms of select arginine tRNAs interact with CNOT3 and promote its recruitment whereas other tRNA D-arms sterically clash with CNOT3. These effects link codon content to mRNA stability. Thus, in addition to their canonical decoding function, tRNAs directly engage regulatory complexes during translation, a mechanism we term P-site tRNA-mediated mRNA decay.
Conflict of interest statement
Figures



Similar articles
-
The Ccr4-Not complex monitors the translating ribosome for codon optimality.Science. 2020 Apr 17;368(6488):eaay6912. doi: 10.1126/science.aay6912. Science. 2020. PMID: 32299921 Free PMC article.
-
Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4-NOT.Nat Struct Mol Biol. 2023 Sep;30(9):1314-1322. doi: 10.1038/s41594-023-01075-8. Epub 2023 Aug 31. Nat Struct Mol Biol. 2023. PMID: 37653243 Free PMC article.
-
Translation elongation and mRNA stability are coupled through the ribosomal A-site.RNA. 2018 Oct;24(10):1377-1389. doi: 10.1261/rna.066787.118. Epub 2018 Jul 11. RNA. 2018. PMID: 29997263 Free PMC article.
-
Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny.Wiley Interdiscip Rev RNA. 2021 Nov;12(6):e1658. doi: 10.1002/wrna.1658. Epub 2021 May 5. Wiley Interdiscip Rev RNA. 2021. PMID: 33949788 Free PMC article. Review.
-
Synonymous Codons: Choose Wisely for Expression.Trends Genet. 2017 Apr;33(4):283-297. doi: 10.1016/j.tig.2017.02.001. Epub 2017 Mar 12. Trends Genet. 2017. PMID: 28292534 Free PMC article. Review.
References
-
- Gallie DR, The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5, 2108–2116 (1991). - PubMed
-
- Garneau NL, Wilusz J, Wilusz CJ, The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8, 113–126 (2007). - PubMed
-
- Yi H et al., PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol Cell 70, 1081–1088 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

