Proteomics of circulating extracellular vesicles reveals diverse clinical presentations of COVID-19 but fails to identify viral peptides
- PMID: 39569406
- PMCID: PMC11576438
- DOI: 10.3389/fcimb.2024.1442743
Proteomics of circulating extracellular vesicles reveals diverse clinical presentations of COVID-19 but fails to identify viral peptides
Abstract
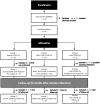
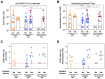
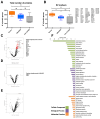
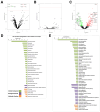
Extracellular vesicles (EVs) released by virus-infected cells have the potential to encapsulate viral peptides, a characteristic that could facilitate vaccine development. Furthermore, plasma-derived EVs may elucidate pathological changes occurring in distal tissues during viral infections. We hypothesized that molecular characterization of EVs isolated from COVID-19 patients would reveal peptides suitable for vaccine development. Blood samples were collected from three cohorts: severe COVID-19 patients (G1), mild/asymptomatic cases (G2), and SARS-CoV-2-negative healthcare workers (G3). Samples were obtained at two time points: during the initial phase of the pandemic in early 2020 (m0) and eight months later (m8). Clinical data analysis revealed elevated inflammatory markers in G1. Notably, non-vaccinated individuals in G1 exhibited increased levels of neutralizing antibodies at m8, suggesting prolonged exposure to viral antigens. Proteomic profiling of EVs was performed using three distinct methods: immunocapture (targeting CD9), ganglioside-capture (utilizing Siglec-1) and size-exclusion chromatography (SEC). Contrary to our hypothesis, this analysis failed to identify viral peptides. These findings were subsequently validated through Western blot analysis targeting the RBD of the SARS-CoV-2 Spike protein's and comparative studies using samples from experimentally infected Syrian hamsters. Furthermore, analysis of the EV cargo revealed a diverse molecular profile, including components involved in the regulation of viral replication, systemic inflammation, antigen presentation, and stress responses. These findings underscore the potential significance of EVs in the pathogenesis and progression of COVID-19.
Keywords: COVID-19 patients; SARS-CoV-2; antibody response; extracellular vesicles; ganglioside-capture (CD169/Siglec-1); immunocapture (CD9); proteomics profiling; size-exclusion chromatography (SEC).
Copyright © 2024 Gualdrón-López, Ayllon-Hermida, Cortes-Serra, Resa-Infante, Bech-Serra, Aparici-Herraiz, Nicolau-Fernandez, Erkizia, Gutierrez-Chamorro, Marfil, Pradenas, Ávila Nieto, Cucurull, Montaner-Tarbés, Muelas, Sotil, Ballana, Urrea, Fraile, Montoya, Vergara, Segales, Carrillo, Izquierdo-Useros, Blanco, Fernandez-Becerra, de La Torre, Pinazo, Martinez-Picado and del Portillo.
Conflict of interest statement
HP, MMo, and LF are shareholders of Innovex Therapeutics. SM-T was a former employee of Innovex Therapeutics. JM-P has received institutional grants and educational/consultancy fees from AbiVax; AstraZeneca; Gilead Sciences; Grifols; Janssen; Merck Sharp & Dohme; and ViiV Healthcare; all outside the submitted work. JC and JB are shareholders of Albajuna Therapeutics SL, NI-U reports institutional grants from Grifols, Dentaid, Hipra and Amassence. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Proteomic profiling of single extracellular vesicles reveals colocalization of SARS-CoV-2 with a CD81/integrin-rich EV subpopulation in sputum from COVID-19 severe patients.Front Immunol. 2023 May 12;14:1052141. doi: 10.3389/fimmu.2023.1052141. eCollection 2023. Front Immunol. 2023. PMID: 37251406 Free PMC article.
-
Immunogenicity and efficacy of XBB.1.5 rS vaccine against the EG.5.1 variant of SARS-CoV-2 in Syrian hamsters.J Virol. 2024 Oct 22;98(10):e0052824. doi: 10.1128/jvi.00528-24. Epub 2024 Sep 4. J Virol. 2024. PMID: 39230305
-
An Intranasal OMV-Based Vaccine Induces High Mucosal and Systemic Protecting Immunity Against a SARS-CoV-2 Infection.Front Immunol. 2021 Dec 17;12:781280. doi: 10.3389/fimmu.2021.781280. eCollection 2021. Front Immunol. 2021. PMID: 34987509 Free PMC article.
-
Emerging roles of extracellular vesicles in COVID-19, a double-edged sword?Immunology. 2021 Aug;163(4):416-430. doi: 10.1111/imm.13329. Epub 2021 May 4. Immunology. 2021. PMID: 33742451 Free PMC article. Review.
-
Potential therapeutic applications of extracellular vesicles in the immunopathogenesis of COVID-19.Pathol Res Pract. 2023 Jan;241:154280. doi: 10.1016/j.prp.2022.154280. Epub 2022 Dec 17. Pathol Res Pract. 2023. PMID: 36580795 Free PMC article. Review.
References
-
- Brustolin M., Rodon J., Rodríguez de la Concepción M. L., Ávila-Nieto C., Cantero G., Pérez M., et al. . (2021). Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 10, 797–809. doi: 10.1080/22221751.2021.1913974 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

