Enhancers and genome conformation provide complex transcriptional control of a herpesviral gene
- PMID: 39562742
- PMCID: PMC11696879
- DOI: 10.1038/s44320-024-00075-0
Enhancers and genome conformation provide complex transcriptional control of a herpesviral gene
Abstract
Complex transcriptional control is a conserved feature of both eukaryotes and the viruses that infect them. Despite viral genomes being smaller and more gene dense than their hosts, we generally lack a sense of scope for the features governing the transcriptional output of individual viral genes. Even having a seemingly simple expression pattern does not imply that a gene's underlying regulation is straightforward. Here, we illustrate this by combining high-density functional genomics, expression profiling, and viral-specific chromosome conformation capture to define with unprecedented detail the transcriptional regulation of a single gene from Kaposi's sarcoma-associated herpesvirus (KSHV). We used as our model KSHV ORF68 - which has simple, early expression kinetics and is essential for viral genome packaging. We first identified seven cis-regulatory regions involved in ORF68 expression by densely tiling the ~154 kb KSHV genome with dCas9 fused to a transcriptional repressor domain (CRISPRi). A parallel Cas9 nuclease screen indicated that three of these regions act as promoters of genes that regulate ORF68. RNA expression profiling demonstrated that three more of these regions act by either repressing or enhancing other distal viral genes involved in ORF68 transcriptional regulation. Finally, we tracked how the 3D structure of the viral genome changes during its lifecycle, revealing that these enhancing regulatory elements are physically closer to their targets when active, and that disrupting some elements caused large-scale changes to the 3D genome. These data enable us to construct a complete model revealing that the mechanistic diversity of this essential regulatory circuit matches that of human genes.
Keywords: CRISPR Interference; Capture Hi-C; Gene Regulation; Herpesvirus; KSHV.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures
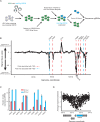
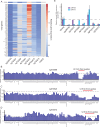
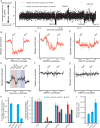
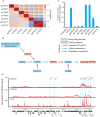


Update of
-
From enhancers to genome conformation: complex transcriptional control underlies expression of a single herpesviral gene.bioRxiv [Preprint]. 2024 Jun 16:2023.07.08.548212. doi: 10.1101/2023.07.08.548212. bioRxiv. 2024. Update in: Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. PMID: 37461644 Free PMC article. Updated. Preprint.
Similar articles
-
From enhancers to genome conformation: complex transcriptional control underlies expression of a single herpesviral gene.bioRxiv [Preprint]. 2024 Jun 16:2023.07.08.548212. doi: 10.1101/2023.07.08.548212. bioRxiv. 2024. Update in: Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. PMID: 37461644 Free PMC article. Updated. Preprint.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The Emerging Roles of Multimolecular G-Quadruplexes in Transcriptional Regulation and Chromatin Organization.Acc Chem Res. 2024 Dec 3;57(23):3397-3406. doi: 10.1021/acs.accounts.4c00574. Epub 2024 Nov 18. Acc Chem Res. 2024. PMID: 39555660 Free PMC article.
-
FoxK1 and FoxK2 cooperate with ORF45 to promote late lytic replication of Kaposi's sarcoma-associated herpesvirus.J Virol. 2024 Dec 17;98(12):e0077924. doi: 10.1128/jvi.00779-24. Epub 2024 Nov 4. J Virol. 2024. PMID: 39494902
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
References
-
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D (2014) KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10:e1003847 - PMC - PubMed
-
- Banerji J, Rusconi S, Schaffner W (1981) Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299–308 - PubMed
MeSH terms
Substances
Grants and funding
- na//Life Sciences Research Foundation (LSRF)
- AI122528/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 1K99AI173531/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- n/a/Howard Hughes Medical Institute (HHMI)
- CA136367/HHS | NIH | National Cancer Institute (NCI)

