Starvation-induced metabolic rewiring affects mTORC1 composition in vivo
- PMID: 39550382
- PMCID: PMC11569187
- DOI: 10.1038/s41598-024-78873-7
Starvation-induced metabolic rewiring affects mTORC1 composition in vivo
Abstract
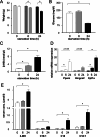
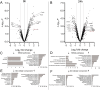
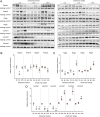
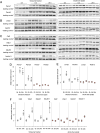
Lysosomes play a crucial role in metabolic adaptation to starvation, but detailed in vivo studies are scarce. Therefore, we investigated the changes of the proteome of liver lysosomes in mice starved short-term for 6h or long-term for 24h. We verified starvation-induced catabolism by weight loss, ketone body production, drop in blood glucose and an increase of 3-methylhistidine. Deactivation of mTORC1 in vivo after short-term starvation causes a depletion of mTORC1 and the associated Ragulator complex in hepatic lysosomes, resulting in diminished phosphorylation of mTORC1 target proteins. While mTORC1 lysosomal protein levels and activity in liver were restored after long-term starvation, the lysosomal levels of Ragulator remained constantly reduced. To determine whether this mTORC1 activity pattern may be organ-specific, we further investigated the key metabolic organs muscle and brain. mTORC1 inactivation, but not re-activation, occurred in muscle after a starvation of 12 h or longer. In brain, mTORC1 activity remained unchanged during starvation. As mTORC1 deactivation is known to induce autophagy, we further investigated the more than 150 non-lysosomal proteins enriched in the lysosomal fraction upon starvation. Proteasomal, cytosolic and peroxisomal proteins dominated after short-term starvation, while after long-term starvation, mainly proteasomal and mitochondrial proteins accumulated, indicating ordered autophagic protein degradation.
© 2024. The Author(s).
Conflict of interest statement
Figures

Similar articles
-
TNFAIP8L2/TIPE2 impairs autolysosome reformation via modulating the RAC1-MTORC1 axis.Autophagy. 2021 Jun;17(6):1410-1425. doi: 10.1080/15548627.2020.1761748. Epub 2020 May 28. Autophagy. 2021. PMID: 32460619 Free PMC article.
-
AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms.Mol Cell Biol. 2018 Apr 30;38(10):e00023-18. doi: 10.1128/MCB.00023-18. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29507183 Free PMC article.
-
PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy.Autophagy. 2017 Mar 4;13(3):486-505. doi: 10.1080/15548627.2016.1263781. Epub 2017 Jan 19. Autophagy. 2017. PMID: 28102733 Free PMC article.
-
mTORC1 and Nutrient Homeostasis: The Central Role of the Lysosome.Int J Mol Sci. 2018 Mar 12;19(3):818. doi: 10.3390/ijms19030818. Int J Mol Sci. 2018. PMID: 29534520 Free PMC article. Review.
-
Regulation of Autophagy through TORC1 and mTORC1.Biomolecules. 2017 Jul 7;7(3):52. doi: 10.3390/biom7030052. Biomolecules. 2017. PMID: 28686223 Free PMC article. Review.
References
-
- Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol21, 101–118. 10.1038/s41580-019-0185-4 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

