Understanding the molecular regulatory mechanisms of autophagy in lung disease pathogenesis
- PMID: 39544928
- PMCID: PMC11560454
- DOI: 10.3389/fimmu.2024.1460023
Understanding the molecular regulatory mechanisms of autophagy in lung disease pathogenesis
Abstract
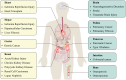
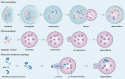
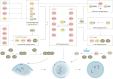
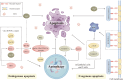
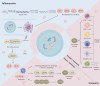
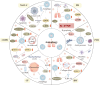
Lung disease development involves multiple cellular processes, including inflammation, cell death, and proliferation. Research increasingly indicates that autophagy and its regulatory proteins can influence inflammation, programmed cell death, cell proliferation, and innate immune responses. Autophagy plays a vital role in the maintenance of homeostasis and the adaptation of eukaryotic cells to stress by enabling the chelation, transport, and degradation of subcellular components, including proteins and organelles. This process is essential for sustaining cellular balance and ensuring the health of the mitochondrial population. Recent studies have begun to explore the connection between autophagy and the development of different lung diseases. This article reviews the latest findings on the molecular regulatory mechanisms of autophagy in lung diseases, with an emphasis on potential targeted therapies for autophagy.
Keywords: COPD; apoptosis; autophagosome; autophagy; pulmonary diseases.
Copyright © 2024 Lin, Lin, Han, Wang, Zhou, Wang, Wang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Valley fever under a changing climate in the United States.Environ Int. 2024 Nov;193:109066. doi: 10.1016/j.envint.2024.109066. Epub 2024 Oct 11. Environ Int. 2024. PMID: 39432997 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

