Post-transcriptional control drives Aurora kinase A expression in human cancers
- PMID: 39527514
- PMCID: PMC11554201
- DOI: 10.1371/journal.pone.0310625
Post-transcriptional control drives Aurora kinase A expression in human cancers
Abstract
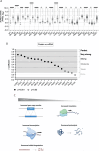
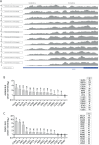
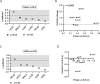
Aurora kinase A (AURKA) is a major regulator of the cell cycle. A prominent association exists between high expression of AURKA and cancer, and impairment of AURKA levels can trigger its oncogenic activity. In order to explore the contribution of post-transcriptional regulation to AURKA expression in different cancers, we carried out a meta-analysis of -omics data of 18 cancer types from The Cancer Genome Atlas (TCGA). Our study confirmed a general trend for increased AURKA mRNA in cancer compared to normal tissues and revealed that AURKA expression is highly dependent on post-transcriptional control in several cancers. Correlation and clustering analyses of AURKA mRNA and protein expression, and expression of AURKA-targeting hsa-let-7a miRNA, unveiled that hsa-let-7a is likely involved to varying extents in controlling AURKA expression in cancers. We then measured differences in the short/long ratio (SLR) of the two alternative cleavage and polyadenylation (APA) isoforms of AURKA mRNA across cancers compared to the respective healthy counterparts. We suggest that the interplay between APA and hsa-let-7a targeting of AURKA mRNA may influence AURKA expression in some cancers. hsa-let-7a and APA may also independently contribute to altered AURKA levels. Therefore, we argue that AURKA mRNA and protein expression are often discordant in cancer as a result of dynamic post-transcriptional regulation.
Copyright: © 2024 Cacioppo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Differential translation of mRNA isoforms underlies oncogenic activation of cell cycle kinase Aurora A.Elife. 2023 Jun 29;12:RP87253. doi: 10.7554/eLife.87253. Elife. 2023. PMID: 37384380 Free PMC article.
-
Regulating the regulator: a survey of mechanisms from transcription to translation controlling expression of mammalian cell cycle kinase Aurora A.Open Biol. 2022 Sep;12(9):220134. doi: 10.1098/rsob.220134. Epub 2022 Sep 7. Open Biol. 2022. PMID: 36067794 Free PMC article. Review.
-
The feedback loop of AURKA/DDX5/TMEM147-AS1/let-7 drives lipophagy to induce cisplatin resistance in epithelial ovarian cancer.Cancer Lett. 2023 Jul 1;565:216241. doi: 10.1016/j.canlet.2023.216241. Epub 2023 May 20. Cancer Lett. 2023. PMID: 37217070
-
SIX3, a tumor suppressor, inhibits astrocytoma tumorigenesis by transcriptional repression of AURKA/B.J Hematol Oncol. 2017 Jun 8;10(1):115. doi: 10.1186/s13045-017-0483-2. J Hematol Oncol. 2017. PMID: 28595628 Free PMC article.
-
Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy.Mol Cancer. 2021 Jan 15;20(1):15. doi: 10.1186/s12943-020-01305-3. Mol Cancer. 2021. PMID: 33451333 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

