Emerging Gene-editing nano-therapeutics for Cancer
- PMID: 39524822
- PMCID: PMC11550658
- DOI: 10.1016/j.heliyon.2024.e39323
Emerging Gene-editing nano-therapeutics for Cancer
Abstract
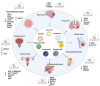
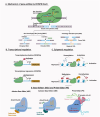
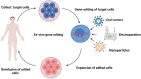
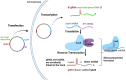
Remarkable progress has been made in the field of genome engineering after the discovery of CRISPR/Cas9 in 2012 by Jennifer Doudna and Emmanuelle Charpentier. Compared to any other gene-editing tools, CRISPR/Cas9 attracted the attention of the scientific community because of its simplicity, specificity, and multiplex editing possibilities for which the inventors were awarded the Nobel prize for chemistry in 2020. CRISPR/Cas9 allows targeted alteration of the genomic sequence, gene regulation, and epigenetic modifications using an RNA-guided site-specific endonuclease. Though the impact of CRISPR/Cas9 was undisputed, some of its limitations led to key modifications including the use of miniature-Cas proteins, Cas9 Retron precise Parallel Editing via homologY (CRISPEY), Cas-Clover, or development of alternative methods including retron-recombineering, Obligate Mobile Element Guided Activity(OMEGA), Fanzor, and Argonaute proteins. As cancer is caused by genetic and epigenetic alterations, gene-editing was found to be highly useful for knocking out oncogenes, editing mutations to regain the normal functioning of tumor suppressor genes, knock-out immune checkpoint blockade in CAR-T cells, producing 'off-the-shelf' CAR-T cells, identify novel tumorigenic genes and functional analysis of multiple pathways in cancer, etc. Advancements in nanoparticle-based delivery of guide-RNA and Cas9 complex to the human body further enhanced the potential of CRISPR/Cas9 for clinical translation. Several studies are reported for developing novel delivery methods to enhance the tumor-specific application of CRISPR/Cas9 for anticancer therapy. In this review, we discuss new developments in novel gene editing techniques and recent progress in nanoparticle-based CRISPR/Cas9 delivery specific to cancer applications.
Keywords: Alternatives of CRISPR/Cas9; CRISPR/Cas9; Cancer; Gene-editing; Nanoparticles; Non-viral delivery.
© 2024 Amrita Vishwa Vidyapeetham Kochi.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Shantikumar Nair reports was provided by Amrita Vishwa Vidyapeetham Amrita Centre for Nanosciences and Molecular Medicine. Shantikumar Nair reports a relationship with Amrita Vishwa Vidyapeetham Amrita Centre for Nanosciences and Molecular Medicine that includes: employment. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer.Mol Cancer. 2021 Oct 1;20(1):126. doi: 10.1186/s12943-021-01431-6. Mol Cancer. 2021. PMID: 34598686 Free PMC article. Review.
-
CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing.Biochemistry (Mosc). 2022 Aug;87(8):777-788. doi: 10.1134/S0006297922080090. Biochemistry (Mosc). 2022. PMID: 36171658 Free PMC article. Review.
-
Harnessing lipid nanoparticles for efficient CRISPR delivery.Biomater Sci. 2021 Sep 14;9(18):6001-6011. doi: 10.1039/d1bm00537e. Biomater Sci. 2021. PMID: 34115079 Free PMC article. Review.
-
Discovery in CRISPR-Cas9 system.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021 Dec 28;46(12):1392-1402. doi: 10.11817/j.issn.1672-7347.2021.210169. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 35232910 Free PMC article. Chinese, English.
-
CRISPR Pioneers Win 2020 Nobel Prize for Chemistry.Iran J Public Health. 2020 Dec;49(12):2235-2239. doi: 10.18502/ijph.v49i12.4800. Iran J Public Health. 2020. PMID: 34178729 Free PMC article.
References
-
- Loeb K.R., Loeb L.A. Significance of multiple mutations in cancer. Carcinogenesis. 2000;21(3):379–385. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

