GPCR-BSD: a database of binding sites of human G-protein coupled receptors under diverse states
- PMID: 39497074
- PMCID: PMC11533411
- DOI: 10.1186/s12859-024-05962-9
GPCR-BSD: a database of binding sites of human G-protein coupled receptors under diverse states
Abstract
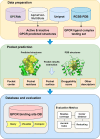
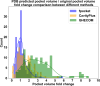
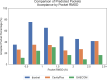
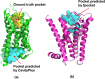
G-protein coupled receptors (GPCRs), the largest family of membrane proteins in human body, involve a great variety of biological processes and thus have become highly valuable drug targets. By binding with ligands (e.g., drugs), GPCRs switch between active and inactive conformational states, thereby performing functions such as signal transmission. The changes in binding pockets under different states are important for a better understanding of drug-target interactions. Therefore it is critical, as well as a practical need, to obtain binding sites in human GPCR structures. We report a database (called GPCR-BSD) that collects 127,990 predicted binding sites of 803 GPCRs under active and inactive states (thus 1,606 structures in total). The binding sites were identified from the predicted GPCR structures by executing three geometric-based pocket prediction methods, fpocket, CavityPlus and GHECOM. The server provides query, visualization, and comparison of the predicted binding sites for both GPCR predicted and experimentally determined structures recorded in PDB. We evaluated the identified pockets of 132 experimentally determined human GPCR structures in terms of pocket residue coverage, pocket center distance and redocking accuracy. The evaluation showed that fpocket and CavityPlus methods performed better and successfully predicted orthosteric binding sites in over 60% of the 132 experimentally determined structures. The GPCR Binding Site database is freely accessible at https://gpcrbs.bigdata.jcmsc.cn . This study not only provides a systematic evaluation of the commonly-used fpocket and CavityPlus methods for the first time but also meets the need for binding site information in GPCR studies.
Keywords: Binding site; Database; G-protein coupled receptor; Key residue; State-specific structure.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Improving the Modeling of Extracellular Ligand Binding Pockets in RosettaGPCR for Conformational Selection.Int J Mol Sci. 2023 Apr 24;24(9):7788. doi: 10.3390/ijms24097788. Int J Mol Sci. 2023. PMID: 37175495 Free PMC article.
-
Multi-state modeling of G-protein coupled receptors at experimental accuracy.Proteins. 2022 Nov;90(11):1873-1885. doi: 10.1002/prot.26382. Epub 2022 May 16. Proteins. 2022. PMID: 35510704 Free PMC article.
-
7TM Domain Structure of Adhesion GPCRs.Handb Exp Pharmacol. 2016;234:43-66. doi: 10.1007/978-3-319-41523-9_3. Handb Exp Pharmacol. 2016. PMID: 27832483 Review.
-
Improving virtual screening of G protein-coupled receptors via ligand-directed modeling.PLoS Comput Biol. 2017 Nov 13;13(11):e1005819. doi: 10.1371/journal.pcbi.1005819. eCollection 2017 Nov. PLoS Comput Biol. 2017. PMID: 29131821 Free PMC article.
-
Conformational changes of G protein-coupled receptors during their activation by agonist binding.J Recept Signal Transduct Res. 2003;23(2-3):123-53. doi: 10.1081/rrs-120025192. J Recept Signal Transduct Res. 2003. PMID: 14626443 Review.
References
-
- Wang J, Hua T, Liu ZJ. Structural features of activated GPCR signaling complexes. Curr Opin Struct Biol. 2020;63:82–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

