Live imaging of airway epithelium reveals that mucociliary clearance modulates SARS-CoV-2 spread
- PMID: 39488529
- PMCID: PMC11531594
- DOI: 10.1038/s41467-024-53791-4
Live imaging of airway epithelium reveals that mucociliary clearance modulates SARS-CoV-2 spread
Abstract
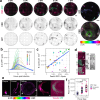
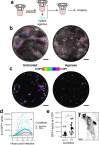
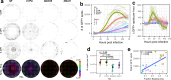
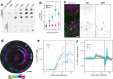
SARS-CoV-2 initiates infection in the conducting airways, where mucociliary clearance inhibits pathogen penetration. However, it is unclear how mucociliary clearance impacts SARS-CoV-2 spread after infection is established. To investigate viral spread at this site, we perform live imaging of SARS-CoV-2 infected differentiated primary human bronchial epithelium cultures for up to 12 days. Using a fluorescent reporter virus and markers for cilia and mucus, we longitudinally monitor mucus motion, ciliary motion, and infection. Infected cell numbers peak at 4 days post infection, forming characteristic foci that tracked mucus movement. Inhibition of MCC using physical and genetic perturbations limits foci. Later in infection, mucociliary clearance deteriorates. Increased mucus secretion accompanies ciliary motion defects, but mucociliary clearance and vectorial infection spread resume after mucus removal, suggesting that mucus secretion may mediate MCC deterioration. Our work shows that while MCC can facilitate SARS-CoV-2 spread after initial infection, subsequent MCC decreases inhibit spread, revealing a complex interplay between SARS-CoV-2 and MCC.
© 2024. The Author(s).
Conflict of interest statement
J.F.H. has received research support, paid to Northwestern University, from Gilead Sciences and is a paid consultant for Merck. All other authors declare no conflicts of interest.
Figures





Update of
-
Live imaging of the airway epithelium reveals that mucociliary clearance modulates SARS-CoV-2 spread.Res Sq [Preprint]. 2023 Sep 8:rs.3.rs-3246773. doi: 10.21203/rs.3.rs-3246773/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Nov 2;15(1):9480. doi: 10.1038/s41467-024-53791-4. PMID: 37720034 Free PMC article. Updated. Preprint.
Similar articles
-
Live imaging of the airway epithelium reveals that mucociliary clearance modulates SARS-CoV-2 spread.Res Sq [Preprint]. 2023 Sep 8:rs.3.rs-3246773. doi: 10.21203/rs.3.rs-3246773/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Nov 2;15(1):9480. doi: 10.1038/s41467-024-53791-4. PMID: 37720034 Free PMC article. Updated. Preprint.
-
Continuous mucociliary transport by primary human airway epithelial cells in vitro.Am J Physiol Lung Cell Mol Physiol. 2015 Jul 15;309(2):L99-108. doi: 10.1152/ajplung.00024.2015. Epub 2015 May 15. Am J Physiol Lung Cell Mol Physiol. 2015. PMID: 25979076 Free PMC article.
-
Long-Term Modeling of SARS-CoV-2 Infection of In Vitro Cultured Polarized Human Airway Epithelium.mBio. 2020 Nov 6;11(6):e02852-20. doi: 10.1128/mBio.02852-20. mBio. 2020. PMID: 33158999 Free PMC article.
-
[Mucociliary clearance in COVID-19 - an underestimated danger area in early phase of disease?].MMW Fortschr Med. 2021 Sep;163(Suppl 5):21-27. doi: 10.1007/s15006-021-0189-9. MMW Fortschr Med. 2021. PMID: 34383284 Free PMC article. Review. German.
-
Role of cilia, mucus, and airway surface liquid in mucociliary dysfunction: lessons from mouse models.J Aerosol Med Pulm Drug Deliv. 2008 Mar;21(1):13-24. doi: 10.1089/jamp.2007.0659. J Aerosol Med Pulm Drug Deliv. 2008. PMID: 18518828 Review.
References
-
- Killingley, B. et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med.28, 1031–1041 (2022). - PubMed
MeSH terms
Grants and funding
- AI094595-09S1/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI118610/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U19 AI135972/AI/NIAID NIH HHS/United States
- AI135964/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI135972/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

