Serum proteomics for the identification of biomarkers to flag predilection of COVID19 patients to various organ morbidities
- PMID: 39487396
- PMCID: PMC11531188
- DOI: 10.1186/s12014-024-09512-6
Serum proteomics for the identification of biomarkers to flag predilection of COVID19 patients to various organ morbidities
Abstract
Background: COVID19 is a pandemic that has affected millions around the world since March 2020. While many patients recovered completely with mild illness, many patients succumbed to various organ morbidities. This heterogeneity in the clinical presentation of COVID19 infection has posed a challenge to clinicians around the world. It is therefore crucial to identify specific organ-related morbidity for effective treatment and better patient outcomes. We have carried out serum-based proteomic experiments to identify protein biomarkers that can flag organ dysfunctions in COVID19 patients.
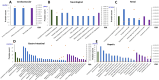
Methods: COVID19 patients were screened and tested at various hospitals across New Delhi, India. 114 serum samples from these patients, with and without organ morbidities were collected and annotated based on clinical presentation and treatment history. Of these, 29 samples comprising of heart, lung, kidney, gastrointestinal, liver, and neurological morbidities were considered for the discovery phase of the experiment. Proteins were isolated, quantified, trypsin digested, and the peptides were subjected to liquid chromatography assisted tandem mass spectrometry analysis. Data analysis was carried out using Proteome Discoverer software. Fold change analysis was carried out on MetaboAnalyst. KEGG, Reactome, and Wiki Pathway analysis of differentially expressed proteins were carried out using the STRING database. Potential biomarker candidates for various organ morbidities were validated using ELISA.
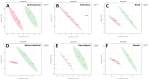
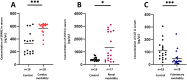
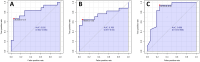
Results: 254 unique proteins were identified from all the samples with a subset of 12-31 differentially expressed proteins in each of the clinical phenotypes. These proteins establish complement and coagulation cascade pathways in the pathogenesis of the organ morbidities. Validation experiments along with their diagnostic parameters confirm Secreted Protein Acidic and Rich in Cysteine, Cystatin C, and Catalase as potential biomarker candidates that can flag cardiovascular disease, renal disease, and respiratory disease, respectively.
Conclusions: Label free serum proteomics shows differential protein expression in COVID19 patients with morbidity as compared to those without morbidity. Identified biomarker candidates hold promise to flag organ morbidities in COVID19 for efficient patient care.
Keywords: Biomarkers; COVID19; Clinical proteomics; Coagulation factors; Complement factors; Differential protein expression; Organ morbidity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Identification of potential biomarkers to predict organ morbidity in COVID-19: A repository based proteomics perspective.Biochem Biophys Rep. 2023 Sep;35:101493. doi: 10.1016/j.bbrep.2023.101493. Epub 2023 Jun 2. Biochem Biophys Rep. 2023. PMID: 37304132 Free PMC article.
-
Quantitative proteomic analysis of human plasma using tandem mass tags to identify novel biomarkers for herpes zoster.J Proteomics. 2020 Aug 15;225:103879. doi: 10.1016/j.jprot.2020.103879. Epub 2020 Jun 30. J Proteomics. 2020. PMID: 32585426
-
Label-free plasma proteomics for the identification of the putative biomarkers of oral squamous cell carcinoma.J Proteomics. 2022 May 15;259:104541. doi: 10.1016/j.jprot.2022.104541. Epub 2022 Feb 26. J Proteomics. 2022. PMID: 35231661
-
Secretome Proteomic Approaches for Biomarker Discovery: An Update on Colorectal Cancer.Medicina (Kaunas). 2020 Aug 31;56(9):443. doi: 10.3390/medicina56090443. Medicina (Kaunas). 2020. PMID: 32878319 Free PMC article. Review.
-
Qualification and Verification of Protein Biomarker Candidates.Adv Exp Med Biol. 2016;919:493-514. doi: 10.1007/978-3-319-41448-5_23. Adv Exp Med Biol. 2016. PMID: 27975232 Review.
References
LinkOut - more resources
Full Text Sources
