Dynamic composition of stress granules in Trypanosoma brucei
- PMID: 39480887
- PMCID: PMC11556693
- DOI: 10.1371/journal.ppat.1012666
Dynamic composition of stress granules in Trypanosoma brucei
Abstract
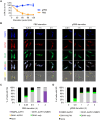
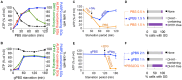
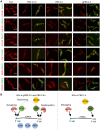
Stress granules (SGs) are stress-induced RNA condensates consisting of stalled initiation complexes resulting from translational inhibition. The biochemical composition and function of SGs are highly diverse, and this diversity has been attributed to different stress conditions, signalling pathways involved and specific cell types. Interestingly, mRNA decay components, which are found in ubiquitous cytoplasmic foci known as processing bodies (PB), have also been identified in SG proteomes. A major challenge in current SG studies is to understand the cause of SG diversity, as well as the function of SG under different stress conditions. Trypanosoma brucei is a single-cellular parasite that causes Human African Trypanosomiasis (sleeping sickness). In this study, we showed that by varying the supply of extracellular carbon sources during starvation, cellular ATP levels changed rapidly, resulting in SGs of different compositions and dynamics. We identified a subset of SG components, which dissociated from the SGs in response to cellular ATP depletion. Using expansion microscopy, we observed sub-granular compartmentalization of PB- and SG-components within the stress granules. Our results highlight the importance of cellular ATP in SG composition and dynamics, providing functional insight to SGs formed under different stress conditions.
Copyright: © 2024 Aye et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Trypanosoma brucei PRMT1 Is a Nucleic Acid Binding Protein with a Role in Energy Metabolism and the Starvation Stress Response.mBio. 2018 Dec 18;9(6):e02430-18. doi: 10.1128/mBio.02430-18. mBio. 2018. PMID: 30563898 Free PMC article.
-
Stress-specific differences in assembly and composition of stress granules and related foci.J Cell Sci. 2017 Mar 1;130(5):927-937. doi: 10.1242/jcs.199240. Epub 2017 Jan 17. J Cell Sci. 2017. PMID: 28096475 Free PMC article.
-
Composition and function of stress granules and P-bodies in plants.Semin Cell Dev Biol. 2024 Mar 15;156:167-175. doi: 10.1016/j.semcdb.2022.11.008. Epub 2022 Dec 1. Semin Cell Dev Biol. 2024. PMID: 36464613 Review.
-
Methods to Classify Cytoplasmic Foci as Mammalian Stress Granules.J Vis Exp. 2017 May 12;(123):55656. doi: 10.3791/55656. J Vis Exp. 2017. PMID: 28570526 Free PMC article.
-
Properties of Stress Granule and P-Body Proteomes.Mol Cell. 2019 Oct 17;76(2):286-294. doi: 10.1016/j.molcel.2019.09.014. Mol Cell. 2019. PMID: 31626750 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

