Rhizospheric miRNAs affect the plant microbiota
- PMID: 39474459
- PMCID: PMC11520407
- DOI: 10.1093/ismeco/ycae120
Rhizospheric miRNAs affect the plant microbiota
Erratum in
-
Correction to: Rhizospheric miRNAs affect the plant microbiota.ISME Commun. 2024 Dec 4;4(1):ycae144. doi: 10.1093/ismeco/ycae144. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39735891 Free PMC article.
Abstract
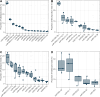
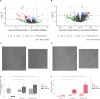
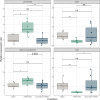
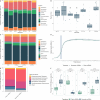
Small ribonucleic acids (RNAs) have been shown to play important roles in cross-kingdom communication, notably in plant-pathogen relationships. Plant micro RNAs (miRNAs)-one class of small RNAs-were even shown to regulate gene expression in the gut microbiota. Plant miRNAs could also affect the rhizosphere microbiota. Here we looked for plant miRNAs in the rhizosphere of model plants, and if these miRNAs could affect the rhizosphere microbiota. We first show that plant miRNAs were present in the rhizosphere of Arabidopsis thaliana and Brachypodium distachyon. These plant miRNAs were also found in or on bacteria extracted from the rhizosphere. We then looked at the effect these plants miRNAs could have on two typical rhizosphere bacteria, Variovorax paradoxus and Bacillus mycoides. The two bacteria took up a fluorescent synthetic miRNA but only V. paradoxus shifted its transcriptome when confronted to a mixture of six plant miRNAs. V. paradoxus also changed its transcriptome when it was grown in the rhizosphere of Arabidopsis that overexpressed a miRNA in its roots. As there were differences in the response of the two isolates used, we looked for shifts in the larger microbial community. We observed shifts in the rhizosphere bacterial communities of Arabidopsis mutants that were impaired in their small RNA pathways, or overexpressed specific miRNAs. We also found differences in the growth and community composition of a simplified soil microbial community when exposed in vitro to a mixture of plant miRNAs. Our results support the addition of miRNAs to the plant tools shaping rhizosphere microbial assembly.
Keywords: Variovorax; bacterial communities; plant miRNAs; rhizosphere; transcriptomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors report no conflict of interest.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
-
Interventions to increase patient and family involvement in escalation of care for acute life-threatening illness in community health and hospital settings.Cochrane Database Syst Rev. 2020 Dec 8;12(12):CD012829. doi: 10.1002/14651858.CD012829.pub2. Cochrane Database Syst Rev. 2020. PMID: 33285618 Free PMC article.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

