DLK1 and DLK2, two non-canonical ligands of NOTCH receptors, differentially modulate the osteogenic differentiation of mesenchymal C3H10T1/2 cells
- PMID: 39473022
- PMCID: PMC11523663
- DOI: 10.1186/s40659-024-00561-7
DLK1 and DLK2, two non-canonical ligands of NOTCH receptors, differentially modulate the osteogenic differentiation of mesenchymal C3H10T1/2 cells
Abstract
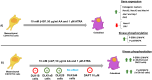
Background: C3H10T1/2 is a mesenchymal cell line capable of differentiating into osteoblasts, adipocytes and chondrocytes. The differentiation of these cells into osteoblasts is modulated by various transcription factors, such as RUNX2. Additionally, several interconnected signaling pathways, including the NOTCH pathway, play a crucial role in modulating their differentiation into mature bone cells. We have investigated the roles of DLK1 and DLK2, two non-canonical inhibitory ligands of NOTCH receptors, in the osteogenic differentiation of C3H10T1/2 cells.
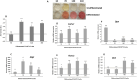
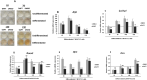
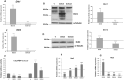
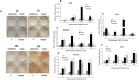
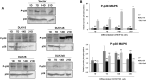
Results: Our results corroborate existing evidence that DLK1 acts as an inhibitor of osteogenesis. In contrast, we demonstrate for the first time that DLK2 enhances this differentiation process. Additionally, our data suggest that NOTCH2, 3 and 4 receptors may promote osteogenesis, as indicated by their increased expression during this process, whereas NOTCH1 expression, which decreases during cell differentiation, might inhibit osteogenesis. Moreover, treatment with DAPT, a NOTCH signaling inhibitor, impeded osteogenic differentiation. We have confirmed the increase in ERK1/2 MAPK and p38 MAPK phosphorylation in C3H10T1/2 cells induced to differentiate to osteoblasts. Our new findings reveal increased ERK1/2 MAPK phosphorylation in differentiated C3H10T1/2 cells with a decrease in DLK1 expression or an overexpression of DLK2, which is coincident with the behavior of those transfectants where we have detected an increase in osteogenic differentiation. Additionally, p38 MAPK phosphorylation increases in differentiated C3H10T1/2 cells with reduced DLK1 levels.
Conclusions: Our results suggest that DLK1 may inhibit osteogenesis, while DLK2 may promote it, by modulating NOTCH signaling and the phosphorylation of ERK1/2 and p38 MAPK pathways. Given the established inhibitory effect of DLK proteins on NOTCH signaling, these new insights could pave the way for developing future therapeutic strategies aimed at treating bone diseases.
Keywords: DLK; ERK1/2 MAPK; Mesenchymal C3H10T1/2 cells; NOTCH; Osteogenesis; p38 MAPK.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other's activities.Biochim Biophys Acta. 2011 Jun;1813(6):1153-64. doi: 10.1016/j.bbamcr.2011.03.004. Epub 2011 Mar 15. Biochim Biophys Acta. 2011. PMID: 21419176
-
NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells.Cells. 2020 Sep 4;9(9):2032. doi: 10.3390/cells9092032. Cells. 2020. PMID: 32899774 Free PMC article.
-
Different Expression Levels of DLK2 Inhibit NOTCH Signaling and Inversely Modulate MDA-MB-231 Breast Cancer Tumor Growth In Vivo.Int J Mol Sci. 2022 Jan 29;23(3):1554. doi: 10.3390/ijms23031554. Int J Mol Sci. 2022. PMID: 35163478 Free PMC article.
-
The imprinted gene Delta like non-canonical Notch ligand 1 (Dlk1) is conserved in mammals, and serves a growth modulatory role during tissue development and regeneration through Notch dependent and independent mechanisms.Cytokine Growth Factor Rev. 2019 Apr;46:17-27. doi: 10.1016/j.cytogfr.2019.03.006. Epub 2019 Mar 23. Cytokine Growth Factor Rev. 2019. PMID: 30930082 Review.
-
Possible roles of DLK1 in the Notch pathway during development and disease.Biochim Biophys Acta. 2012 Jun;1822(6):988-95. doi: 10.1016/j.bbadis.2012.02.003. Epub 2012 Feb 12. Biochim Biophys Acta. 2012. PMID: 22353464 Review.
References
-
- Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–16. - PubMed
-
- Lai EC. Notch cleavage: Nicastrin helps Presenilin make the final cut. Curr Biol. 2002;12(6):R200–2. - PubMed
-
- Barrick D, Kopan R. The Notch transcription activation complex makes its move. Cell. 2006;124(5):883–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

