Discovery of Novel Inhibitors against ALS-Related SOD1(A4V) Aggregation through the Screening of a Chemical Library Using Differential Scanning Fluorimetry (DSF)
- PMID: 39458929
- PMCID: PMC11510448
- DOI: 10.3390/ph17101286
Discovery of Novel Inhibitors against ALS-Related SOD1(A4V) Aggregation through the Screening of a Chemical Library Using Differential Scanning Fluorimetry (DSF)
Abstract
Background: Cu/Zn Superoxide Dismutase 1 (SOD1) is a 32 kDa cytosolic dimeric metalloenzyme that neutralizes superoxide anions into oxygen and hydrogen peroxide. Mutations in SOD1 are associated with ALS, a disease causing motor neuron atrophy and subsequent mortality. These mutations exert their harmful effects through a gain of function mechanism, rather than a loss of function. Despite extensive research, the mechanism causing selective motor neuron death still remains unclear. A defining feature of ALS pathogenesis is protein misfolding and aggregation, evidenced by ubiquitinated protein inclusions containing SOD1 in affected motor neurons. This work aims to identify compounds countering SOD1(A4V) misfolding and aggregation, which could potentially aid in ALS treatment.
Methods: The approach employed was in vitro screening of a library comprising 1280 pharmacologically active compounds (LOPAC®) in the context of drug repurposing. Using differential scanning fluorimetry (DSF), these compounds were tested for their impact on SOD1(A4V) thermal stability.
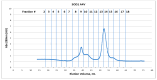
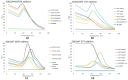
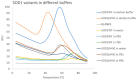
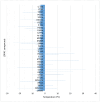
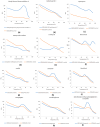
Results and conclusions: Dimer stability was the parameter chosen as the criterion for screening, since the dissociation of the native SOD1 dimer is the step prior to its in vitro aggregation. The screening revealed one compound raising protein-ligand Tm by 6 °C, eleven inducing a higher second Tm, suggesting a stabilization effect, and fourteen reducing Tm from 10 up to 26 °C, suggesting possible interactions or non-specific binding.
Keywords: ALS; DSF technique; SOD1; chemical library scanning; drug discovery; drug repurposing; protein-misfolding diseases.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Quercitrin and quercetin 3-β-d-glucoside as chemical chaperones for the A4V SOD1 ALS-causing mutant.Protein Eng Des Sel. 2017 Jun 1;30(6):431-440. doi: 10.1093/protein/gzx025. Protein Eng Des Sel. 2017. PMID: 28475686 Free PMC article.
-
Cupric ions induce the oxidation and trigger the aggregation of human superoxide dismutase 1.PLoS One. 2013 Jun 3;8(6):e65287. doi: 10.1371/journal.pone.0065287. Print 2013. PLoS One. 2013. PMID: 23755211 Free PMC article.
-
Mutant Cu/Zn Superoxide Dismutase (A4V) Turnover Is Altered in Cells Containing Inclusions.Front Mol Neurosci. 2021 Nov 3;14:771911. doi: 10.3389/fnmol.2021.771911. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34803609 Free PMC article.
-
Hydrogen Peroxide and Amyotrophic Lateral Sclerosis: From Biochemistry to Pathophysiology.Antioxidants (Basel). 2021 Dec 27;11(1):52. doi: 10.3390/antiox11010052. Antioxidants (Basel). 2021. PMID: 35052556 Free PMC article. Review.
-
Does wild-type Cu/Zn-superoxide dismutase have pathogenic roles in amyotrophic lateral sclerosis?Transl Neurodegener. 2020 Aug 19;9(1):33. doi: 10.1186/s40035-020-00209-y. Transl Neurodegener. 2020. PMID: 32811540 Free PMC article. Review.
References
-
- Mehta P.R., Jones A.R., Opie-Martin S., Shatunov A., Iacoangeli A., Al Khleifat A., Smith B.N., Topp S., Morrison K.E., Shaw P.J., et al. Younger age of onset in familial amyotrophic lateral sclerosis is a result of pathogenic gene variants, rather than ascertainment bias. J. Neurol. Neurosurg. Psychiatry. 2019;90:268–271. doi: 10.1136/jnnp-2018-319089. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

