In Silico Discovery of a Novel PI3Kδ Inhibitor Incorporating 3,5,7-Trihydroxychroman-4-one Targeting Diffuse Large B-Cell Lymphoma
- PMID: 39457034
- PMCID: PMC11508633
- DOI: 10.3390/ijms252011250
In Silico Discovery of a Novel PI3Kδ Inhibitor Incorporating 3,5,7-Trihydroxychroman-4-one Targeting Diffuse Large B-Cell Lymphoma
Abstract
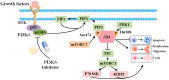
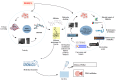
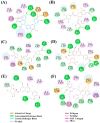
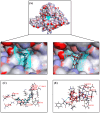
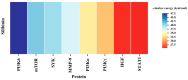
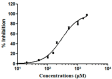
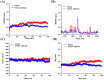
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoma, and it is highly aggressive and heterogeneous. Targeted therapy is still the main treatment method used in clinic due to its lower risk of side effects and personalized medication. Excessive activation of PI3Kδ in DLBCL leads to abnormal activation of the PI3K/Akt pathway, promoting the occurrence and development of DLBCL. The side effects of existing PI3Kδ inhibitors limit their clinical application. The discovery of PI3Kδ inhibitors with novel structures and minimal side effects is urgently needed. This study constructed a PI3Kδ inhibitor screening model to screen natural product libraries. Revealing the mechanism of natural product therapy for DLBCL through network pharmacology, kinase assays, and molecular dynamics. The results of molecular docking indicated that Silibinin had a high docking score and a good binding mode with PI3Kδ. The results of network pharmacology indicated that Silibinin could exert therapeutic effects on DLBCL by inhibiting PI3Kδ activity and affecting the PI3K/Akt pathway. The kinase assays indicated that Silibinin concentration dependently inhibited the activity of PI3Kδ. The results of molecular dynamics indicated that Silibinin could stably bind to PI3Kδ. Silibinin was a structurally novel 3,5,7-trihydroxychroman-4-one PI3Kδ inhibitor, providing valuable information for the subsequent discovery of PI3Kδ inhibitors.
Keywords: DLBCL; PI3Kδ; molecular dynamics; network pharmacology; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest in this article.
Figures



Similar articles
-
SAF-248, a novel PI3Kδ-selective inhibitor, potently suppresses the growth of diffuse large B-cell lymphoma.Acta Pharmacol Sin. 2022 Jan;43(1):209-219. doi: 10.1038/s41401-021-00644-1. Epub 2021 Mar 29. Acta Pharmacol Sin. 2022. PMID: 33782541 Free PMC article.
-
Identification of novel PI3Kδ inhibitors by docking, ADMET prediction and molecular dynamics simulations.Comput Biol Chem. 2019 Feb;78:190-204. doi: 10.1016/j.compbiolchem.2018.12.002. Epub 2018 Dec 7. Comput Biol Chem. 2019. PMID: 30557817
-
Discovery of novel quinazoline derivatives as potent PI3Kδ inhibitors with high selectivity.Eur J Med Chem. 2020 Dec 15;208:112865. doi: 10.1016/j.ejmech.2020.112865. Epub 2020 Sep 21. Eur J Med Chem. 2020. PMID: 32987316
-
A Review of Phosphocreatine 3 Kinase δ Subtype (PI3Kδ) and Its Inhibitors in Malignancy.Med Sci Monit. 2021 Oct 9;27:e932772. doi: 10.12659/MSM.932772. Med Sci Monit. 2021. PMID: 34625526 Free PMC article. Review.
-
Inhibition of the PI3K/Akt/mTOR signaling pathway in diffuse large B-cell lymphoma: current knowledge and clinical significance.Molecules. 2014 Sep 11;19(9):14304-15. doi: 10.3390/molecules190914304. Molecules. 2014. PMID: 25215588 Free PMC article. Review.
References
-
- Uddin S., Hussain A.R., Siraj A.K., Manogaran P.S., Al-Jomah N.A., Moorji A., Atizado V., Al-Dayel F., Belgaumi A., El-Solh H., et al. Role of phosphatidylinositol 3’-kinase/AKT pathway in diffuse large B-cell lymphoma survival. Blood. 2006;108:4178–4186. doi: 10.1182/blood-2006-04-016907. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

