FUS::DDIT3 Fusion Protein in the Development of Myxoid Liposarcoma and Possible Implications for Therapy
- PMID: 39456230
- PMCID: PMC11506083
- DOI: 10.3390/biom14101297
FUS::DDIT3 Fusion Protein in the Development of Myxoid Liposarcoma and Possible Implications for Therapy
Abstract
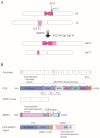
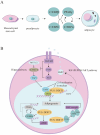
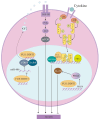
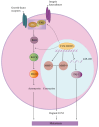
The FUS::DDIT3 fusion protein, formed by the chromosomal translocation t (12;16) (q13;p11), is found in over 90% of myxoid liposarcoma (MLS) cases and is a crucial protein in its development. Many studies have explored the role of FUS::DDIT3 in MLS, and the prevailing view is that FUS::DDIT3 inhibits adipocyte differentiation and promotes MLS growth and invasive migration by functioning as an aberrant transcription factor that affects gene expression and regulates its downstream molecules. As fusion proteins are gradually showing their potential as targets for precision cancer therapy, FUS::DDIT3 has also been investigated as a therapeutic target. Drugs that target FUS::DDIT3 and its downstream molecules for treating MLS are widely utilized in both clinical practice and experimental studies, and some of them have demonstrated promising results. This article reviews the findings of relevant research, providing an overview of the oncogenic mechanisms of the FUS::DDIT3 fusion protein in MLS, as well as recent advancements in its therapy.
Keywords: FUS::DDIT3 fusion protein; chromosomal translocation; myxoid liposarcoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Exploiting WEE1 Kinase Activity as FUS::DDIT3-Dependent Therapeutic Vulnerability in Myxoid Liposarcoma.Clin Cancer Res. 2024 Nov 1;30(21):4974-4986. doi: 10.1158/1078-0432.CCR-24-1152. Clin Cancer Res. 2024. PMID: 39207225
-
Nuclear expression of FLT1 and its ligand PGF in FUS-DDIT3 carrying myxoid liposarcomas suggests the existence of an intracrine signaling loop.BMC Cancer. 2010 Jun 1;10:249. doi: 10.1186/1471-2407-10-249. BMC Cancer. 2010. PMID: 20515481 Free PMC article.
-
The myxoid/round cell liposarcoma fusion oncogene FUS-DDIT3 and the normal DDIT3 induce a liposarcoma phenotype in transfected human fibrosarcoma cells.Am J Pathol. 2006 May;168(5):1642-53. doi: 10.2353/ajpath.2006.050872. Am J Pathol. 2006. PMID: 16651630 Free PMC article.
-
Understanding mesenchymal cancer: the liposarcoma-associated FUS-DDIT3 fusion gene as a model.Semin Cancer Biol. 2005 Jun;15(3):206-14. doi: 10.1016/j.semcancer.2005.01.006. Semin Cancer Biol. 2005. PMID: 15826835 Review.
-
Gene of the month: DDIT3.J Clin Pathol. 2024 Mar 20;77(4):211-216. doi: 10.1136/jcp-2023-208963. J Clin Pathol. 2024. PMID: 38053287 Review.
References
-
- International Agency for Research on Cancer . Soft Tissue and Bone Tumours. International Agency for Research on Cancer; Lyon, France: 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

