Skeletal muscle reprogramming enhances reinnervation after peripheral nerve injury
- PMID: 39455585
- PMCID: PMC11511891
- DOI: 10.1038/s41467-024-53276-4
Skeletal muscle reprogramming enhances reinnervation after peripheral nerve injury
Abstract
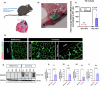
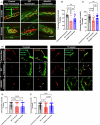
Peripheral Nerve Injuries (PNI) affect more than 20 million Americans and severely impact quality of life by causing long-term disability. PNI is characterized by nerve degeneration distal to the site of nerve injury resulting in long periods of skeletal muscle denervation. During this period, muscle fibers atrophy and frequently become incapable of "accepting" innervation because of the slow speed of axon regeneration post injury. We hypothesize that reprogramming the skeletal muscle to an embryonic-like state may preserve its reinnervation capability following PNI. To this end, we generate a mouse model in which NANOG, a pluripotency-associated transcription factor is expressed locally upon delivery of doxycycline (Dox) in a polymeric vehicle. NANOG expression in the muscle upregulates the percentage of Pax7+ nuclei and expression of eMYHC along with other genes that are involved in muscle development. In a sciatic nerve transection model, NANOG expression leads to upregulation of key genes associated with myogenesis, neurogenesis and neuromuscular junction (NMJ) formation. Further, NANOG mice demonstrate extensive overlap between synaptic vesicles and NMJ acetylcholine receptors (AChRs) indicating restored innervation. Indeed, NANOG mice show greater improvement in motor function as compared to wild-type (WT) animals, as evidenced by improved toe-spread reflex, EMG responses and isometric force production. In conclusion, we demonstrate that reprogramming muscle can be an effective strategy to improve reinnervation and functional outcomes after PNI.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Skeletal muscle reprogramming enhances reinnervation after peripheral nerve injury.Res Sq [Preprint]. 2024 Jan 5:rs.3.rs-3463557. doi: 10.21203/rs.3.rs-3463557/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Oct 25;15(1):9218. doi: 10.1038/s41467-024-53276-4 PMID: 38260278 Free PMC article. Updated. Preprint.
Similar articles
-
Skeletal muscle reprogramming enhances reinnervation after peripheral nerve injury.Res Sq [Preprint]. 2024 Jan 5:rs.3.rs-3463557. doi: 10.21203/rs.3.rs-3463557/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Oct 25;15(1):9218. doi: 10.1038/s41467-024-53276-4 PMID: 38260278 Free PMC article. Updated. Preprint.
-
Macrophage-Derived Vascular Endothelial Growth Factor-A Is Integral to Neuromuscular Junction Reinnervation after Nerve Injury.J Neurosci. 2020 Dec 9;40(50):9602-9616. doi: 10.1523/JNEUROSCI.1736-20.2020. Epub 2020 Nov 6. J Neurosci. 2020. PMID: 33158964 Free PMC article.
-
Terminal Schwann cells participate in neuromuscular synapse remodeling during reinnervation following nerve injury.J Neurosci. 2014 Apr 30;34(18):6323-33. doi: 10.1523/JNEUROSCI.4673-13.2014. J Neurosci. 2014. PMID: 24790203 Free PMC article.
-
Applications of Proteomics to Nerve Regeneration Research.In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 15. In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 15. PMID: 21882439 Free Books & Documents. Review.
-
Chapter 25: Phototherapy in peripheral nerve injury: effects on muscle preservation and nerve regeneration.Int Rev Neurobiol. 2009;87:445-64. doi: 10.1016/S0074-7742(09)87025-6. Int Rev Neurobiol. 2009. PMID: 19682654 Review.
References
-
- Taylor, C. A., Braza, D., Rice, J. B. & Dillingham, T. The incidence of peripheral nerve injury in extremity trauma. Am. J. Phys. Med. Rehab.87, 381–385 (2008). - PubMed
-
- Rivera, J. C., Glebus, G. & Cho, M. Disability following combat-sustained nerve injury of the upper limb. bone Jt. J.96, 254–258 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

