Silybin Meglumine Mitigates CCl4-Induced Liver Fibrosis and Bile Acid Metabolism Alterations
- PMID: 39452937
- PMCID: PMC11509150
- DOI: 10.3390/metabo14100556
Silybin Meglumine Mitigates CCl4-Induced Liver Fibrosis and Bile Acid Metabolism Alterations
Abstract
Background: Altered patterns of bile acids (BAs) are frequently present in liver fibrosis, and BAs function as signaling molecules to initiate inflammatory responses. Silybin meglumine (SLB-M) is widely used in treating various liver diseases including liver fibrosis. However, research on its effects on bile acid (BA) metabolism is limited. This study investigated the therapeutic effects of SLB-M on liver fibrosis and BA metabolism in a CCl4-induced murine model.
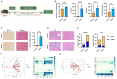
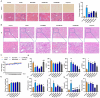
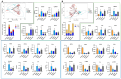
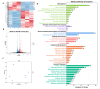
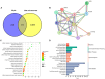
Methods: A murine liver fibrosis model was induced by CCl4. Fibrosis was evaluated using HE, picrosirius red, and Masson's trichrome staining. Liver function was assessed by serum and hepatic biochemical markers. Bile acid (BA) metabolism was analyzed using LC-MS/MS. Bioinformatics analyses, including PPI network, GO, and KEGG pathway analyses, were employed to explore molecular mechanisms. Gene expression alterations in liver tissue were examined via qRT-PCR.
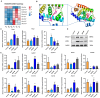
Results: SLB-M treatment resulted in significant histological improvements in liver tissue, reducing collagen deposition and restoring liver architecture. Biochemically, SLB-M not only normalized serum liver enzyme levels (ALT, AST, TBA, and GGT) but also mitigated disruptions in both systemic and hepatic BA metabolism by increased unconjugated BAs like cholic acid and chenodeoxycholic acid but decreased conjugated BAs including taurocholic acid and taurodeoxycholic acid, compared to that in CCl4-induced murine model. Notably, SLB-M efficiently improved the imbalance of BA homeostasis in liver caused by CCl4 via activating Farnesoid X receptor.
Conclusions: These findings underscore SLB-M decreased inflammatory response, reconstructed BA homeostasis possibly by regulating key pathways, and gene expressions in BA metabolism.
Keywords: bile acid; bioinformatics; carbon tetrachloride; liver fibrosis; silybin meglumine.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Si-Wu-Tang ameliorates fibrotic liver injury via modulating intestinal microbiota and bile acid homeostasis.Chin Med. 2021 Nov 4;16(1):112. doi: 10.1186/s13020-021-00524-0. Chin Med. 2021. PMID: 34736501 Free PMC article.
-
Transcriptomic analysis reveals pharmacological mechanisms mediating efficacy of Yangyinghuoxue Decoction in CCl4-induced hepatic fibrosis in rats.Front Pharmacol. 2024 May 15;15:1364023. doi: 10.3389/fphar.2024.1364023. eCollection 2024. Front Pharmacol. 2024. PMID: 38813108 Free PMC article.
-
Hepatoprotective effect of phillygenin on carbon tetrachloride-induced liver fibrosis and its effects on short chain fatty acid and bile acid metabolism.J Ethnopharmacol. 2022 Oct 5;296:115478. doi: 10.1016/j.jep.2022.115478. Epub 2022 Jun 16. J Ethnopharmacol. 2022. PMID: 35716920
-
Hepatoprotective effect of total flavonoids of Mallotus apelta (Lour.) Muell.Arg. leaf against carbon tetrachloride-induced liver fibrosis in rats via modulation of TGF-β1/Smad and NF-κB signaling pathways.J Ethnopharmacol. 2020 May 23;254:112714. doi: 10.1016/j.jep.2020.112714. Epub 2020 Feb 24. J Ethnopharmacol. 2020. PMID: 32105750
-
Pegbelfermin selectively reduces secondary bile acid concentrations in patients with non-alcoholic steatohepatitis.JHEP Rep. 2021 Nov 12;4(1):100392. doi: 10.1016/j.jhepr.2021.100392. eCollection 2022 Jan. JHEP Rep. 2021. PMID: 34977519 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

