Non-Nucleoside Lycorine-Based Analogs as Potential DENV/ZIKV NS5 Dual Inhibitors: Structure-Based Virtual Screening and Chemoinformatic Analysis
- PMID: 39452899
- PMCID: PMC11509260
- DOI: 10.3390/metabo14100519
Non-Nucleoside Lycorine-Based Analogs as Potential DENV/ZIKV NS5 Dual Inhibitors: Structure-Based Virtual Screening and Chemoinformatic Analysis
Abstract
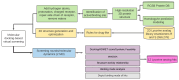
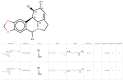
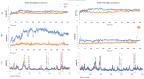
Dengue (DENV) and Zika (ZIKV) virus continue to pose significant challenges globally due to their widespread prevalence and severe health implications. Given the absence of effective vaccines and specific therapeutics, targeting the highly conserved NS5 RNA-dependent RNA polymerase (RdRp) domain has emerged as a promising strategy. However, limited efforts have been made to develop inhibitors for this crucial target. In this study, we employed an integrated in silico approach utilizing combinatorial chemistry, docking, molecular dynamics simulations, MM/GBSA, and ADMET studies to target the allosteric N-pocket of DENV3-RdRp and ZIKV-RdRp. Using this methodology, we designed lycorine analogs with natural S-enantiomers (LYCS) and R-enantiomers (LYCR) as potential inhibitors of non-structural protein 5 (NS5) in DENV3 and ZIKV. Notably, 12 lycorine analogs displayed a robust binding free energy (<-9.00 kcal/mol), surpassing that of RdRp-ribavirin (<-7.00 kcal/mol) along with promising ADMET score predictions (<4.00), of which (LYCR728-210, LYCS728-210, LYCR728-212, LYCS505-214) displayed binding properties to both DENV3 and ZIKV targets. Our research highlights the potential of non-nucleoside lycorine-based analogs with different enantiomers that may present different or even completely opposite metabolic, toxicological, and pharmacological profiles as promising candidates for inhibiting NS5-RdRp in ZIKV and DENV3, paving the way for further exploration for the development of effective antiviral agents.
Keywords: DENV3; MD simulations; MM/GBSA; NS5; ZIKV; compound library; lycorine; molecular docking.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.J Virol. 2020 Oct 14;94(21):e00794-20. doi: 10.1128/JVI.00794-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796069 Free PMC article.
-
Pharmacophore-Model-Based Drug Repurposing for the Identification of the Potential Inhibitors Targeting the Allosteric Site in Dengue Virus NS5 RNA-Dependent RNA Polymerase.Viruses. 2022 Aug 20;14(8):1827. doi: 10.3390/v14081827. Viruses. 2022. PMID: 36016449 Free PMC article.
-
NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues.J Virol. 2019 Dec 12;94(1):e01294-19. doi: 10.1128/JVI.01294-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597763 Free PMC article.
-
The Flavivirus Non-Structural Protein 5 (NS5): Structure, Functions, and Targeting for Development of Vaccines and Therapeutics.Vaccines (Basel). 2024 Aug 1;12(8):865. doi: 10.3390/vaccines12080865. Vaccines (Basel). 2024. PMID: 39203991 Free PMC article. Review.
-
Perspectives on the current antiviral developments towards RNA-dependent RNA polymerase (RdRp) and methyltransferase (MTase) domains of dengue virus non-structural protein 5 (DENV-NS5).Eur J Med Chem. 2023 Aug 5;256:115416. doi: 10.1016/j.ejmech.2023.115416. Epub 2023 Apr 28. Eur J Med Chem. 2023. PMID: 37159959 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

