Comprehensive RNA-Seq Gene Co-Expression Analysis Reveals Consistent Molecular Pathways in Hepatocellular Carcinoma across Diverse Risk Factors
- PMID: 39452074
- PMCID: PMC11505157
- DOI: 10.3390/biology13100765
Comprehensive RNA-Seq Gene Co-Expression Analysis Reveals Consistent Molecular Pathways in Hepatocellular Carcinoma across Diverse Risk Factors
Abstract
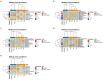
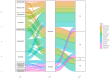
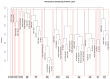
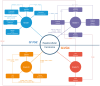
Hepatocellular carcinoma (HCC) has the highest mortality rate and is the most frequent of liver cancers. The heterogeneity of HCC in its etiology and molecular expression increases the difficulty in identifying possible treatments. To elucidate the molecular mechanisms of HCC across grades, data from The Cancer Genome Atlas (TCGA) were used for gene co-expression analysis, categorizing each sample into its pre-existing risk factors. The R library BioNERO was used for preprocessing and gene co-expression network construction. For those modules most correlated with a grade, functional enrichments from different databases were then tested, which appeared to have relatively consistent patterns when grouped by G1/G2 and G3/G4. G1/G2 exhibited the involvement of pathways related to metabolism and the PI3K/Akt pathway, which regulates cell proliferation and related pathways, whereas G3/G4 showed the activation of cell adhesion genes and the p53 signaling pathway, which regulates apoptosis, cell cycle arrest, and similar processes. Module preservation analysis was then used with the no history dataset as the reference network, which found cell adhesion molecules and cell cycle genes to be preserved across all risk factors, suggesting they are imperative in the development of HCC regardless of potential etiology. Through hierarchical clustering, modules related to the cell cycle, cell adhesion, the immune system, and the ribosome were found to be consistently present across all risk factors, with distinct clusters linked to oxidative phosphorylation in viral HCC and pentose and glucuronate interconversions in non-viral HCC, underscoring their potential roles in cancer progression.
Keywords: cancer etiology; hepatocellular carcinoma; histological grades; module enrichment; module preservation; pathway analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Transcriptome Analysis Revealed a Highly Connected Gene Module Associated With Cirrhosis to Hepatocellular Carcinoma Development.Front Genet. 2019 Apr 2;10:305. doi: 10.3389/fgene.2019.00305. eCollection 2019. Front Genet. 2019. PMID: 31001331 Free PMC article.
-
Identification and validation of real hub genes in hepatocellular carcinoma based on weighted gene co-expression network analysis.Cancer Biomark. 2022;35(2):227-243. doi: 10.3233/CBM-220151. Cancer Biomark. 2022. PMID: 36120772
-
Silencing of long noncoding RNA HOXD-AS1 inhibits proliferation, cell cycle progression, migration and invasion of hepatocellular carcinoma cells through MEK/ERK pathway.J Cell Biochem. 2020 Jan;121(1):443-457. doi: 10.1002/jcb.29206. Epub 2019 Jun 23. J Cell Biochem. 2020. PMID: 31231887
-
Upregulation of Stress-Induced Protein Kinase CK1 Delta is associated with a Poor Prognosis for patients with Hepatocellular Carcinoma.Genet Test Mol Biomarkers. 2021 Jul;25(7):504-514. doi: 10.1089/gtmb.2020.0093. Genet Test Mol Biomarkers. 2021. PMID: 34280005
-
Aloperine induces apoptosis and G2/M cell cycle arrest in hepatocellular carcinoma cells through the PI3K/Akt signaling pathway.Phytomedicine. 2019 Aug;61:152843. doi: 10.1016/j.phymed.2019.152843. Epub 2019 Jan 28. Phytomedicine. 2019. PMID: 31039533
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

