Comparative analysis of methodologies for detecting extrachromosomal circular DNA
- PMID: 39448595
- PMCID: PMC11502736
- DOI: 10.1038/s41467-024-53496-8
Comparative analysis of methodologies for detecting extrachromosomal circular DNA
Abstract
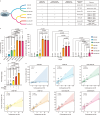
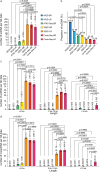
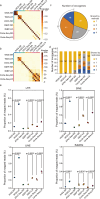
Extrachromosomal circular DNA (eccDNA) is crucial in oncogene amplification, gene transcription regulation, and intratumor heterogeneity. While various analysis pipelines and experimental methods have been developed for eccDNA identification, their detection efficiencies have not been systematically assessed. To address this, we evaluate the performance of 7 analysis pipelines using seven simulated datasets, in terms of accuracy, identity, duplication rate, and computational resource consumption. We also compare the eccDNA detection efficiency of 7 experimental methods through twenty-one real sequencing datasets. Here, we show that Circle-Map and Circle_finder (bwa-mem-samblaster) outperform the other short-read pipelines. However, Circle_finder (bwa-mem-samblaster) exhibits notable redundancy in its outcomes. CReSIL is the most effective pipeline for eccDNA detection in long-read sequencing data at depths higher than 10X. Moreover, long-read sequencing-based Circle-Seq shows superior efficiency in detecting copy number-amplified eccDNA over 10 kb in length. These results offer valuable insights for researchers in choosing the suitable methods for eccDNA research.
© 2024. The Author(s).
Conflict of interest statement
Jingwen Fang is the chief executive officer of HanGen Biotech. The other authors declare no competing interests.
Figures

Similar articles
-
Bioinformatics advances in eccDNA identification and analysis.Oncogene. 2024 Oct;43(41):3021-3036. doi: 10.1038/s41388-024-03138-6. Epub 2024 Aug 29. Oncogene. 2024. PMID: 39209966 Review.
-
CReSIL: accurate identification of extrachromosomal circular DNA from long-read sequences.Brief Bioinform. 2022 Nov 19;23(6):bbac422. doi: 10.1093/bib/bbac422. Brief Bioinform. 2022. PMID: 36198068 Free PMC article.
-
ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data.BMC Bioinformatics. 2022 Jan 14;23(1):40. doi: 10.1186/s12859-021-04545-2. BMC Bioinformatics. 2022. PMID: 35030991 Free PMC article.
-
Skeletal Muscles of Sedentary and Physically Active Aged People Have Distinctive Genic Extrachromosomal Circular DNA Profiles.Int J Mol Sci. 2023 Feb 1;24(3):2736. doi: 10.3390/ijms24032736. Int J Mol Sci. 2023. PMID: 36769072 Free PMC article.
-
Extrachromosomal circular DNA in cancer: history, current knowledge, and methods.Trends Genet. 2022 Jul;38(7):766-781. doi: 10.1016/j.tig.2022.02.007. Epub 2022 Mar 8. Trends Genet. 2022. PMID: 35277298 Review.
References
-
- Cox, D., Yuncken, C. & Spriggs, A. I. Minute chromatin bodies in malignant tumours of childhood. Lancet1, 55–58 (1965). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

