PCNA-binding activity separates RNF168 functions in DNA replication and DNA double-stranded break signaling
- PMID: 39445802
- PMCID: PMC11602139
- DOI: 10.1093/nar/gkae918
PCNA-binding activity separates RNF168 functions in DNA replication and DNA double-stranded break signaling
Abstract
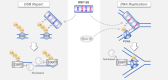
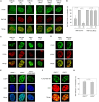
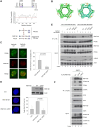
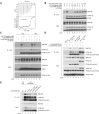
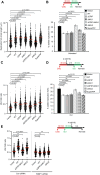
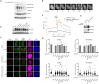
RNF168 orchestrates a ubiquitin-dependent DNA damage response to regulate the recruitment of repair factors, such as 53BP1 to DNA double-strand breaks (DSBs). In addition to its canonical functions in DSB signaling, RNF168 may facilitate DNA replication fork progression. However, the precise role of RNF168 in DNA replication remains unclear. Here, we demonstrate that RNF168 is recruited to DNA replication factories in a manner that is independent of the canonical DSB response pathway regulated by Ataxia-Telangiectasia Mutated (ATM) and RNF8. We identify a degenerate Proliferating Cell Nuclear Antigen (PCNA)-interacting peptide (DPIP) motif in the C-terminus of RNF168, which together with its Motif Interacting with Ubiquitin (MIU) domain mediates binding to mono-ubiquitylated PCNA at replication factories. An RNF168 mutant harboring inactivating substitutions in its DPIP box and MIU1 domain (termed RNF168 ΔDPIP/ΔMIU1) is not recruited to sites of DNA synthesis and fails to support ongoing DNA replication. Notably, the PCNA interaction-deficient RNF168 ΔDPIP/ΔMIU1 mutant fully rescues the ability of RNF168-/- cells to form 53BP1 foci in response to DNA DSBs. Therefore, RNF168 functions in DNA replication and DSB signaling are fully separable. Our results define a new mechanism by which RNF168 promotes DNA replication independently of its canonical functions in DSB signaling.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
53BP1-the 'Pandora's box' of genome integrity.DNA Repair (Amst). 2024 Dec;144:103779. doi: 10.1016/j.dnarep.2024.103779. Epub 2024 Oct 28. DNA Repair (Amst). 2024. PMID: 39476547 Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase.Science. 2007 Dec 7;318(5856):1637-40. doi: 10.1126/science.1150034. Epub 2007 Nov 15. Science. 2007. PMID: 18006705 Free PMC article.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Continuing education meetings and workshops: effects on professional practice and healthcare outcomes.Cochrane Database Syst Rev. 2021 Sep 15;9(9):CD003030. doi: 10.1002/14651858.CD003030.pub3. Cochrane Database Syst Rev. 2021. PMID: 34523128 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

