Engineering a cleaved, prefusion-stabilized influenza B virus hemagglutinin by identification and locking of all six pH switches
- PMID: 39445049
- PMCID: PMC11497598
- DOI: 10.1093/pnasnexus/pgae462
Engineering a cleaved, prefusion-stabilized influenza B virus hemagglutinin by identification and locking of all six pH switches
Abstract
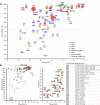
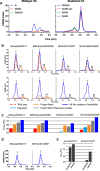
Vaccine components based on viral fusion proteins require high stability of the native prefusion conformation for optimal potency and manufacturability. In the case of influenza B virus hemagglutinin (HA), the stem's conformation relies on efficient cleavage. In this study, we identified six pH-sensitive regions distributed across the entire ectodomain where protonated histidines assume either a repulsive or an attractive role. Substitutions in these areas enhanced the protein's expression, quality, and stability in its prefusion trimeric state. Importantly, this stabilization enabled the production of a cleavable HA0, which is further processed into HA1 and HA2 by furin during exocytic pathway passage, thereby facilitating correct folding, increased stability, and screening for additional stabilizing substitutions in the core of the metastable fusion domain. Cryo-EM analysis at neutral and low pH revealed a previously unnoticed pH switch involving the C-terminal residues of the natively cleaved HA1. This switch keeps the fusion peptide in a clamped state at neutral pH, averting premature conformational shift. Our findings shed light on new strategies for possible improvements of recombinant or genetic-based influenza B vaccines.
© The Author(s) 2024. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures



Similar articles
-
Intermonomer Interactions in Hemagglutinin Subunits HA1 and HA2 Affecting Hemagglutinin Stability and Influenza Virus Infectivity.J Virol. 2015 Oct;89(20):10602-11. doi: 10.1128/JVI.00939-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269180 Free PMC article.
-
Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2208011119. doi: 10.1073/pnas.2208011119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939703 Free PMC article.
-
Universal stabilization of the influenza hemagglutinin by structure-based redesign of the pH switch regions.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2115379119. doi: 10.1073/pnas.2115379119. Proc Natl Acad Sci U S A. 2022. PMID: 35131851 Free PMC article.
-
Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin.Annu Rev Biochem. 2000;69:531-69. doi: 10.1146/annurev.biochem.69.1.531. Annu Rev Biochem. 2000. PMID: 10966468 Review.
-
Early steps of the conformational change of influenza virus hemagglutinin to a fusion active state: stability and energetics of the hemagglutinin.Biochim Biophys Acta. 2003 Jul 11;1614(1):3-13. doi: 10.1016/s0005-2736(03)00158-5. Biochim Biophys Acta. 2003. PMID: 12873761 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

