A comprehensive framework for trans-ancestry pathway analysis using GWAS summary data from diverse populations
- PMID: 39441834
- PMCID: PMC11534268
- DOI: 10.1371/journal.pgen.1011322
A comprehensive framework for trans-ancestry pathway analysis using GWAS summary data from diverse populations
Abstract
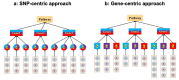
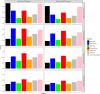
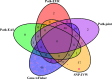
As more multi-ancestry GWAS summary data become available, we have developed a comprehensive trans-ancestry pathway analysis framework that effectively utilizes this diverse genetic information. Within this framework, we evaluated various strategies for integrating genetic data at different levels-SNP, gene, and pathway-from multiple ancestry groups. Through extensive simulation studies, we have identified robust strategies that demonstrate superior performance across diverse scenarios. Applying these methods, we analyzed 6,970 pathways for their association with schizophrenia, incorporating data from African, East Asian, and European populations. Our analysis identified over 200 pathways significantly associated with schizophrenia, even after excluding genes near genome-wide significant loci. This approach substantially enhances detection efficiency compared to traditional single-ancestry pathway analysis and the conventional approach that amalgamates single-ancestry pathway analysis results across different ancestry groups. Our framework provides a flexible and effective tool for leveraging the expanding pool of multi-ancestry GWAS summary data, thereby improving our ability to identify biologically relevant pathways that contribute to disease susceptibility.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Ceftazidime with avibactam for treating severe aerobic Gram-negative bacterial infections: technology evaluation to inform a novel subscription-style payment model.Health Technol Assess. 2024 Oct;28(73):1-230. doi: 10.3310/YAPL9347. Health Technol Assess. 2024. PMID: 39487661 Free PMC article.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The complex genetic architecture of Alzheimer's disease: novel insights and future directions.EBioMedicine. 2023 Apr;90:104511. doi: 10.1016/j.ebiom.2023.104511. Epub 2023 Mar 10. EBioMedicine. 2023. PMID: 36907103 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

