Molecular mechanisms underlying glucose-dependent insulinotropic polypeptide secretion in human duodenal organoids
- PMID: 39441374
- PMCID: PMC11663192
- DOI: 10.1007/s00125-024-06293-3
Molecular mechanisms underlying glucose-dependent insulinotropic polypeptide secretion in human duodenal organoids
Abstract
Aims/hypothesis: Glucose-dependent insulinotropic polypeptide (GIP) is an incretin hormone secreted by enteroendocrine K cells in the proximal small intestine. This study aimed to explore the function of human K cells at the molecular and cellular levels.
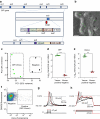
Methods: CRISPR-Cas9 homology-directed repair was used to insert transgenes encoding a yellow fluorescent protein (Venus) or an Epac-based cAMP sensor (Epac-S-H187) in the GIP locus in human duodenal-derived organoids. Fluorescently labelled K cells were purified by FACS for RNA-seq and peptidomic analysis. GIP reporter organoids were employed for GIP secretion assays, live-cell imaging of Ca2+ using Fura-2 and cAMP using Epac-S-H187, and basic electrophysiological characterisation. The G protein-coupled receptor genes GPR142 and CASR were knocked out to evaluate roles in amino acid sensing.
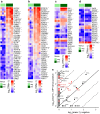
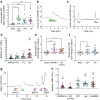
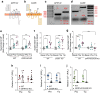
Results: RNA-seq of human duodenal K cells revealed enrichment of several G protein-coupled receptors involved in nutrient sensing, including FFAR1, GPBAR1, GPR119, CASR and GPR142. Glucose induced action potential firing and cytosolic Ca2+ elevation and caused a 1.8-fold increase in GIP secretion, which was inhibited by the sodium glucose co-transporter 1/2 (SGLT1/2) blocker sotagliflozin. Activation of the long-chain fatty acid receptor free fatty acid receptor 1 (FFAR1) induced a 2.7-fold increase in GIP secretion, while tryptophan and phenylalanine stimulated secretion by 2.8- and 2.1-fold, respectively. While CASR knockout blunted intracellular Ca2+ responses, a CASR/GPR142 double knockout was needed to reduce GIP secretory responses to aromatic amino acids.
Conclusions/interpretation: The newly generated human organoid K cell model enables transcriptomic and functional characterisation of nutrient-sensing pathways involved in human GIP secretion. Both calcium-sensing receptor (CASR) and G protein-coupled receptor 142 (GPR142) contribute to protein-stimulated GIP secretion. This model will be further used to identify potential targets for modulation of native GIP secretion in diabetes and obesity.
Keywords: CASR; GIP; GPR142; Organoid; SGLT1.
© 2024. The Author(s).
Conflict of interest statement
Acknowledgements: We thank the MRL Genomics and Transcriptomics Core, the Core Biochemical Assay Laboratory (CBAL), the Flow Cytometry Core at CIMR, the CRUK Cambridge Institute Genomics Core and Addenbrooke’s Tissue Bank. Some of the data were presented as an abstract at the 58th EASD Annual Meeting of the EASD in 2022 and the 5th European Incretin Study Group meeting in 2024. Data availability: RNA-seq data are deposited in the National Center for Biotechnology Information–Gene Expression Omnibus (NCBI GEO) repository (GSE271017). Mass spectrometry proteomics data are deposited to the ProteomeXchange Consortium via the PRIDE partner repository (PXD052659). Funding: This research was funded by a Wellcome joint investigator award to FR/FMG (220271/Z/20/Z) and the MRC-Metabolic Diseases Unit (MRC_MC_UU_12012/3). NG was funded by an MRC studentship. Core support was provided by the MRC (MRC_MC_UU_00014/5) and Wellcome (100574/Z/12/Z). The LC-MS/MS instrument was funded by the MRC (MR/M009041/1). Authors’ relationships and activities: FMG and FR received funding from AstraZeneca and Eli Lilly for non-overlapping research on other projects. They received sponsorship from AstraZeneca, Eli Lilly, Sun Pharma and Mercodia to run the 5th European Incretin Study Group conference in Cambridge (April 2024). The authors declare that there are no other relationships or activities that might bias, or be perceived to bias, their work. Contribution statement: NG generated the majority of the results and wrote the first draft of the manuscript. CA and AD did the electrophysiology. ELM, MS-H and RBB provided training and support, including for organoid maintenance, CRISPR and live-cell imaging, helping with initial data collection and analysis. RGK performed and analysed LC-MS/MS. CAS performed bioinformatics analysis of RNA-seq. FR and FMG designed and oversaw the study, revised the manuscript and are guarantors of the work. All authors contributed to manuscript revision and approved the published version.
Figures

Similar articles
-
Single-cell transcriptomics of human organoid-derived enteroendocrine cell populations from the small intestine.J Physiol. 2024 Dec 5:10.1113/JP287463. doi: 10.1113/JP287463. Online ahead of print. J Physiol. 2024. PMID: 39639676 Free PMC article.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Activation of Gs signaling in mouse enteroendocrine K cells greatly improves obesity- and diabetes-related metabolic deficits.J Clin Invest. 2024 Oct 22;134(24):e182325. doi: 10.1172/JCI182325. J Clin Invest. 2024. PMID: 39436694 Free PMC article.
-
Cellular mechanisms governing glucose-dependent insulinotropic polypeptide secretion.Peptides. 2020 Mar;125:170206. doi: 10.1016/j.peptides.2019.170206. Epub 2019 Nov 19. Peptides. 2020. PMID: 31756367 Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
References
-
- Guccio N, Gribble FM, Reimann F (2022) Glucose-dependent insulinotropic polypeptide-a postprandial hormone with unharnessed metabolic potential. Annu Rev Nutr 42:21–44. 10.1146/annurev-nutr-062320-113625 - PubMed
-
- O’Neil PM, Birkenfeld AL, McGowan B et al (2018) Efficacy and safety of semaglutide compared with liraglutide and placebo for weight loss in patients with obesity: a randomised, double-blind, placebo and active controlled, dose-ranging, phase 2 trial. Lancet 392(10148):637–649. 10.1016/S0140-6736(18)31773-2 - PubMed
-
- Nauck MA, Heimesaat MM, Orskov C, Holst JJ, Ebert R, Creutzfeldt W (1993) Preserved incretin activity of glucagon-like peptide 1 [7-36 amide] but not of synthetic human gastric inhibitory polypeptide in patients with type-2 diabetes mellitus. J Clin Invest 91(1):301–307. 10.1172/JCI116186 - PMC - PubMed
-
- Jastreboff AM, Aronne LJ, Ahmad NN et al (2022) Tirzepatide once weekly for the treatment of obesity. N Engl J Med 387(3):205–216. 10.1056/NEJMoa2206038 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

